Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Similares em
SciELO
Similares em
SciELO
Compartilhar
Revista mexicana de ciencias pecuarias
versão On-line ISSN 2448-6698versão impressa ISSN 2007-1124
Rev. mex. de cienc. pecuarias vol.11 no.4 Mérida Out./Dez. 2020 Epub 02-Mar-2021
https://doi.org/10.22319/rmcp.v11i4.5302
Articles
Genomic diversity and structure of Lidia breed cattle in Mexico
a Universidad Complutense de Madrid. Facultad de Veterinaria, Departamento de Producción Animal. Avenida Puerta de Hierro, s/n, 28040, Madrid, España.
First documented in the 13th Century on the Iberian Peninsula, the Lidia cattle breed has since been the preferred breed for producing bulls for social celebrations known as “bullfighting”, an expression of regional cultural identity in several countries. Specialization of the breed in Mexico began in the late 19th Century when four Mexican families imported a small number of Lidia animals from Spain. Of these original imports, only the lines derived from the Llaguno and González families remain. Different breeding strategies were implemented in the Llaguno family. Antonio Llaguno crossed the recently imported Spanish animals among each other, resulting in what is currently recognized in Mexico as the “Pure” line. Julián Llaguno crossed Creole dams with Spanish sires, creating the line known as “Impure”. In addition, Lidia breed lines such as Domecq, Murube and Santa Coloma were brought to Mexico between 1996 and 1997. The present study objective was to use SNP molecular markers to analyze genomic diversity, population structure, endogamy levels and genetic relationships between Lidia lines in Mexico. Five lines within the Mexican population were studied: Antonio Llaguno, Julián Llaguno, González, Domecq and Santa Coloma. All five lines were found to be genetically distinct, although the Antonio and Julián Llaguno lines are more similar than the others. Genetic isolation between the different lines of the Lidia breed in Mexico has resulted in their being unique.
Key words Lidia breed; population genetics; genetic diversity; genetic structure
La raza bovina de Lidia ha sido seleccionada desde el siglo XIII para participar en festejos sociales reconocidos bajo el término de “Tauromaquia”. En la actualidad forman parte de la identidad de las culturas regionales de varios países. En México, la raza se especializó a finales del siglo XIX cuando cuatro familias mexicanas importaron un número reducido de bovinos de España. De estas importaciones actualmente solo permanecen las líneas derivadas de las familias Llaguno y González. En la familia Llaguno se llevaron diferentes estrategias de reproducción. Antonio Llaguno cruzó los recién importados bovinos españoles entre sí; de dichas cruzas derivó la línea que actualmente es reconocida como “Pura”. Por otro lado, Julián Llaguno realizó cruzas entre hembras criollas con machos españoles, línea conocida como “Impura”. Por último, entre1996 y 1997, un grupo de ganaderos importó bovinos pertenecientes a ciertos encastes españoles como Domecq, Murube, Santa Coloma, entre otros. El objetivo del presente estudio fue investigar la diversidad genómica, estructura poblacional, niveles de endogamia y relaciones genéticas entre las poblaciones de Lidia mexicana, utilizando marcadores moleculares de tipo SNP. La población fue dividida en cinco grupos: Antonio Llaguno, Julián Llaguno, González, y dos grupos que incluían bien importaciones recientes de origen Domecq, o bien importaciones recientes de origen Santa Coloma. Los resultados permiten apreciar diferentes orígenes genéticos dentro de la familia Llaguno en función de su origen histórico: Antonio y Julián. En el resto de grupos también se observa una clara diferenciación genética. Este aislamiento genético entre poblaciones de Lidia mexicana es una característica de su singularidad.
Palabras clave Raza de Lidia; Genética de poblaciones; Diversidad genética; Estructura genética
Introduction
First documented in the 13th Century on the Iberian Peninsula, the Lidia cattle breed is distinguished by selection for behavioral characteristics that enhance aggressiveness and for its use in civil and religious events1. Various social and cultural phenomena involving bulls are currently included in what is known as “bullfighting”2,3. Several countries consider different bullfighting traditions as practices that reinforce regional cultural identity2,3; indeed, in Spain and Peru it has been designated an intangible cultural heritage4. The Lidia breed is characterized for having low genetic and ecological interchangeability5,6.
The first documented bullfighting celebration in Mexico was held in 1523 using aggressive cattle brought mainly from the Navarra region in Spain, which is where the Casta Navarra breed originates7. It was not until the turn of the 20th Century, however, that specialized breeding of Lidia began in Mexico with importation of a small number of animals from Spain by four breeding families: Llaguno, González, Barbabosa and Madrazo8,9. Only genetic lines originating with the Llaguno and González families are still extant today8,10.
The Llaguno family has been located largely in north-central Mexico. Under Antonio Llaguno the reproduction system was closed involving crosses only between Lidia animals directly linked to the original imported animals; in Mexican livestock terminology these are known as “Pure” animals. Julián Llaguno, brother of Antonio, followed a different breeding strategy, crossing Creole dams with Lidia sires of known Spanish origin; these are termed “Impure”8,9. The González family, located in south-central Mexico, crosses imported Lidia breed animals with local cattle selected for aggressiveness8. With the purpose of breeding bulls for bullfights, in 1996 and 1997 a group of Mexican breeders imported animals from Spanish lines such as Domecq, Murube, Santa Coloma and Saltillo, among others; this strategy ended when livestock imports were prohibited for animal health reasons10.
The current Lidia breed population in Mexico is approximately 110,000 animals raised on a total of around 135,000 ha10. This breed is raised under extensive conditions, which favors conservation of endemic flora and fauna. Its central role in many local social traditions supports Mexico’s livestock economy while reinforcing regional cultural identity7,10,11.
The genetic variability of the Lidia breed population in Mexico versus the original Spanish population has been analyzed using autosomal microsatellite markers, with differentiation between Spanish Lidia lines and the Llaguno and González family lines12. These results were confirmed using molecular data produced with DNA chips for bi-allelic single nucleotide polymorphism (SNP) molecular markers. Clear genetic differentiation has also been reported between the Antonio Llaguno and González family lines13,14, although these analyses did not include samples from the Julián Llaguno line or the lines imported in the late 20th Century (Domecq, Santa Coloma, etc.).
The present study objective was to use SNP molecular markers to analyze the genomic diversity, population structure, endogamy levels and genetic relationships in representative populations of the Lidia breed in Mexico.
Material and methods
A total of 306 blood samples were randomly collected from animals belonging to 32 ranches in Mexico affiliated with the Union of Lidia Bull Breeders (Unión de Criadores de Toros de Lidia). The samples were classified into five lines based on the historical origins of each: Antonio Llaguno, Julián Llaguno, González, Domecq and Santa Coloma (Table 1). The samples were collected in tubes containing Magic Buffer® preservative (Biogen Diagnostica, Spain) and kept at 15 °C until DNA extraction. Genomic DNA was extracted using a standard phenol/chloroform protocol15, and the samples were later genotyped with the 50K medium density SNP bovine chip (http://www.illumina.com).
Table 1 Number of analyzed animals (N), genetic distance by ranch and averaged by line (F ST ), endogamy coefficient (F IS ), observed heterozygosity (Ho) and genetic diversity (He)
| Line | Ranch | N | F ST | F IS | Ho | He |
|---|---|---|---|---|---|---|
| Julian Llaguno | Pozo Hondo | 21 | 0.05 | 0.13 | 0.27 | 0.73 |
| Valparaiso | 15 | 0.06 | 0.19 | 0.25 | 0.75 | |
| El Sauz | 8 | 0.07 | 0.21 | 0.24 | 0.76 | |
| Caparica | 11 | 0.05 | 0.17 | 0.26 | 0.74 | |
| Total | 55 | Avg. = 0.06 | ||||
| Antonio Llaguno | San Mateo | 6 | 0.09 | 0.21 | 0.24 | 0.76 |
| Reyes Huerta | 39 | 0.06 | 0.21 | 0.24 | 0.76 | |
| Fernando de la Mora | 6 | 0.10 | 0.04 | 0.30 | 0.70 | |
| Los Cues | 7 | 0.07 | 0.29 | 0.22 | 0.78 | |
| Garfias | 6 | 0.08 | 0.26 | 0.23 | 0.77 | |
| Antigua | 6 | 0.09 | 0.27 | 0.23 | 0.77 | |
| Xajay | 6 | 0.05 | 0.15 | 0.26 | 0.74 | |
| Teófilo Gómez | 6 | 0.07 | 0.19 | 0.25 | 0.75 | |
| Celia Barbabosa | 6 | 0.06 | 0.15 | 0.26 | 0.74 | |
| Boquilla del Cármen | 6 | 0.06 | 0.29 | 0.22 | 0.78 | |
| Fermín Rivera | 6 | 0.07 | 0.18 | 0.25 | 0.75 | |
| Corlomé | 6 | 0.13 | 0.00 | 0.31 | 0.69 | |
| Arroyo Zarco | 6 | 0.05 | 0.18 | 0.26 | 0.74 | |
| Marrón | 6 | 0.05 | 0.11 | 0.28 | 0.72 | |
| La Punta | 19 | 0.10 | 0.11 | 0.27 | 0.73 | |
| Total | 137 | Avg. = 0.07 | ||||
| González | Tenexac | 8 | 0.13 | 0.29 | 0.22 | 0.78 |
| Yturbe | 5 | 0.11 | 0.14 | 0.27 | 0.73 | |
| De Haro | 6 | 0.08 | 0.12 | 0.27 | 0.73 | |
| Castañeda | 6 | 0.11 | 0.25 | 0.23 | 0.77 | |
| Zacatepec | 12 | 0.12 | 0.03 | 0.30 | 0.70 | |
| Rancho Seco | 6 | 0.14 | 0.05 | 0.29 | 0.71 | |
| Total | 43 | Avg. = 0.11 | ||||
| Domecq | La Joya | 17 | 0.10 | 0.15 | 0.26 | 0.74 |
| Santa Maria de Xalpa | 17 | 0.08 | 0.06 | 0.29 | 0.71 | |
| Jaral de Peñas | 17 | 0.06 | -0.03 | 0.32 | 0.68 | |
| Torreon de Cañas | 6 | 0.09 | -0.02 | 0.32 | 0.68 | |
| Jose Julian Llaguno | 10 | 0.07 | 0.00 | 0.31 | 0.69 | |
| Total | 106 | Avg. = 0.08 | ||||
| Santa Coloma | Los Encinos | 5 | 0.02 | 0.17 | 0.26 | 0.74 |
| San José | 6 | 0.02 | 0.10 | 0.28 | 0.72 | |
| Total | 11 | Avg. = 0.02 |
Using the PLINK ver. 1.07 software16, the information was refined by excluding SNPs located on sex chromosomes, those exhibiting a minor (<0.01) allele frequency (MAF), those with <20% missing genotypes and those diverging from Hardy-Weinberg equilibrium (P<0.001). A total of 41,455 SNPs remained for analysis.
Again using PLINK16, analyses were done of three genetic diversity parameters: observed heterozygosity (Ho), expected heterozygosity (He) and the endogamy coefficient (F IS ), estimated as 1-Ho/He. The F ST coefficients were calculated using the ARLEQUIN ver. 3.0 software17. For each individual, the proportion of genetic origins identifiable using the Bayesian grouping algorithm was calculated with the ADMIXTURE software18,19. Graphs were generated with the POPHELPER ver. 1.0.10 software20.
A molecular analysis of variance (AMOVA) was run using a linear model to evaluate genetic variation between and within lines17. The analysis was done in hierarchical mode with three levels (between lines, between ranches in the same line and within ranches). The same software was used to calculate mean distance of the lines in terms of F ST .
Individual runs of homozygosity (ROHs) were identified per individual21. This was done using PLINK with 30 SNP windows, allowing for <100 kb between two consecutive homozygous SNPs, less than two missing genotypes, one heterozygous and a 500 kbp minimum length. The average value per ranch and per line was then calculated.
Results and discussion
Genetic diversity
Average endogamy (F IS ) values per ranch ranged from -0.03 (Jaral de Peñas) to 0.29 (Los Cues, Boquilla del Cármen and Tenexac) (Table 1). The excess heterozygotes present at the Jaral de Peñas and Torreon de Cañas ranches, both in the Domecq line, explains the negative F IS values as a consequence of the Wallhund effect22. The average F ST distances estimated per line were similar among them (0.06 - 0.11), while average F ST distances estimated per ranch ranged from 0.02 (Los Encinos and San José) to 0.14 (Rancho Seco). Ranchers in Mexico are known to exchange sires and dams, a practice more common among ranchers belonging to the same livestock groups and/or working with the same breed. Exchange frequency and the quantity of animals involved undoubtedly depends on rancher criteria, but this could explain the minimal genetic distances between ranches in the same line.
Of total genetic variability, 10.8% was due to interline differences and 6.9% to differences between ranches in the same line (Table 2). It is to be expected that the lack of interline exchanges generates greater differences between lines than within them, where exchanges occur more regularly. The average interline F ST value observed here (0.18) was similar to the average F ST value reported for the Lidia population in Spain (0.15) but higher than found in other cattle breeds (values near 0.07)6. High F ST values result from the characteristic structure of the Lidia breed, in which subdivision into subpopulations or lines produces small effective group sizes.
Genetic structure and population differentiation
The cross-validation error (CV) used in ADMIXTURE calculates values that decrease as the number of hypothetical ancestral populations (K) increases. When the CV value begins to increase it indicates the most probable hypothetical population prediction. Using the present data the most accurate prediction was identified at K = 518,19.
The average proportions of individuals in the ranches coincided with their assignment to each of the five ancestral populations (Figure 1). Each of the five lines largely corresponded to one of the five defined ancestral populations. Discrimination between the Julián Llaguno and Antonio Llaguno lines was less evident between some ranches in these lines, while it was greater between others (e.g., El Sauz and Valparaíso in Julián Llaguno, and Garfias, Los Cués and La Antigua in Antonio Llaguno). This analysis does not explain these differentiations between the Llaguno lines. Perhaps they result from variation in original genetic material since the Julián Llaguno line includes crosses between Creole dams and Spanish Lidia sires. Nonetheless, it is clear that both line (Antonio Llaguno and Julián Llaguno) mostly share common genetic origins. In contrast, the Gonzáles, Domecq and Santa Coloma lines are clearly genetically distinct.
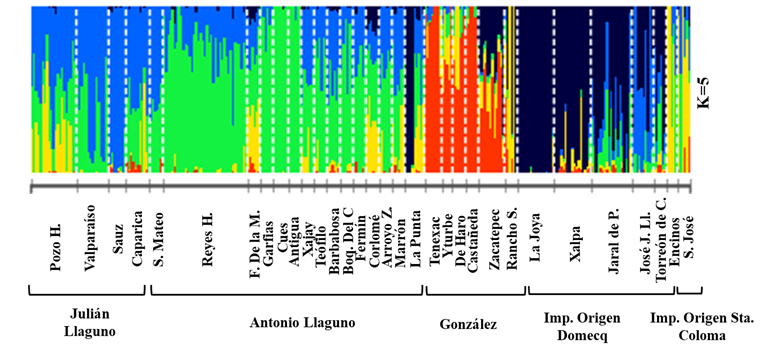
Each vertical line represents an individual animal’s total genome. The proportion of each color (genetic group, K) in the vertical lines is the proportion of each of the five ancestral populations in an individual’s genome (K).
Figure 1 Cross-validation error analysis of hypothetical ancestral populations (K) using ADMIXTURE
Identification of the ROHs per line produced statistics on the average number of runs or segments and average ROH length in each of the five lines (Figure 2). The number and length of ROHs in the different lines exhibited similar patterns. Greater number of segments and length of the ROH are correlated with recent consanguinity events23.
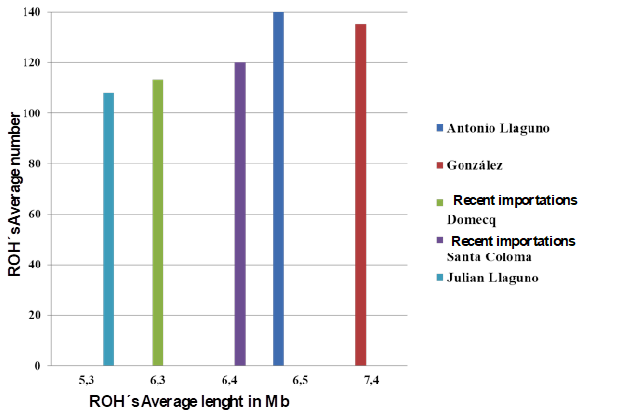
Figure 2 Average number of runs of homozygosity (ROHs) and their average size (Mb) in the five studied lines of the Lidia breed in Mexico
In the data for average ROH number and length by ranch (Table 3), length ranged from 5 (Jaral de Peñas) to 7.7 Mb (Boquilla del Carmen and El Sauz). This is consistent with the average F IS values in which the highest values (>0.20) corresponded to Boquilla del Carmen and El Sauz while the lowest was for Jaral de Peñas (Table 1).
Table 3 Average number of runs of homozygosity (ROH) per ranch, including number of segments (NSEG) and average length (Mb)
| Line | Ranch | NSEG | Average length (Mb) |
|---|---|---|---|
| Julián Llaguno | Pozo Hondo | 108 | 5.3 |
| Valparaiso | 134 | 6.2 | |
| El Sauz | 111 | 7.7 | |
| Caparica | 114 | 6.5 | |
| Pomedio | 117 | 6.1 | |
| Antonio Llaguno | San Mateo | 141 | 6.5 |
| Reyes Huerta | 124 | 5.9 | |
| Fernando de la Mora | 87 | 5.8 | |
| Garfias | 139 | 7.1 | |
| Los Cués | 131 | 7.3 | |
| La Antigua | 144 | 6.9 | |
| Xajay | 115 | 6.6 | |
| Teófilo Gómez | 135 | 6.0 | |
| Celia Barbabosa | 126 | 5.8 | |
| Boquilla del Cármen | 132 | 7.7 | |
| Fermín Rivera | 131 | 6.1 | |
| Corlomé | 79 | 5.3 | |
| Arroyo Zarco | 124 | 6.3 | |
| Marrón | 105 | 6.2 | |
| La Punta | 104 | 6.1 | |
| Pomedio | 121 | 6.2 | |
| González | Tenexac | 135 | 7.4 |
| Gonzalo Yturbe | 110 | 6.3 | |
| De Haro | 113 | 5.6 | |
| C.Castañeda | 134 | 6.9 | |
| Zacatepec | 83 | 5.7 | |
| Rancho Seco | 95 | 5.2 | |
| Pomedio | 109 | 6.2 | |
| Domecq | La Joya | 113 | 6.3 |
| Sta. Maria de Xalpa | 92 | 5.7 | |
| Jaral de Peñas | 64 | 5.0 | |
| José Julián Llaguno | 72 | 5.1 | |
| Torréon de Cañas | 73 | 5.2 | |
| Pomedio | 85 | 5.5 | |
| Santa Coloma | Los Encinos | 120 | 6.4 |
| San José | 105 | 5.8 | |
| Pomedio | 112 | 6.1 |
When animals are isolated in relatively small populations the probability is greater that they inherit identical DNA segments that account for ROH23. In previous studies, the high number and long length of ROHs have been associated with endogamy23. This coincides with the results observed here for the five studied Lidia breed lines in Mexico, which had values greater than those reported in previous analyses of ROHs in native Spanish and American Creole breeds13. Both the ROH and F IS values in the studied Mexican Lidia population reflect subdivision into lines and its consequences: reduction of effective sizes and higher consanguinity values24. This subdivision is effective at preserving intrapopulation genetic variability25,;however, as each subpopulation experiences genetic drift consanguinity will increase and genetic variability will decrease. Under these circumstances it is advisable to closely monitor degrees of endogamy.
Conclusions and implications
The genetic differentiation observed among the Mexican Lidia population, into lines and even between ranches, is due to the different genetic origins of some lines (i.e. Domecq and Santa Coloma) in conjunction with the genetic isolation maintained between the remaining lines (i.e. Antonio Llaguno, Julián Llaguno and González). Both the genetic structure and ROH analyses identified genetic isolation between the lines of the Mexican Lidia population, which has contributed to genomic variations when compared to European Lidia populations. The Mexican Lidia lines are clearly unique from their ancestral populations and quite differentiated amongst themselves.
Acknowledgements
The research reported here was financed by the “Macro-proyecto de Caracterización genética de la Raza de Lidia Mexicana” supported by the Consejo Nacional de los Recursos Genéticos Pecuarios (CONARGEN) and the Asociación de Criadores de Toros de Lidia. Thanks are due the Genetics Laboratory of the Animal Production Department of the Veterinary Faculty, Universidad Complutense de Madrid, and the participating Mexican Lidia ranchers.
REFERENCES
1. Domecq JP. Del toreo a la bravura. España, Alianza Editorial, 2009. [ Links ]
2. Maudet JB. Terres de taureaux: les jeux taurins de L´Europe à L´Amerique. 1ra ed. España, Casa de Velzaquez, 2010. [ Links ]
3. Prieto-Garrido JL. El toro bravo, ganaderías míticas. España Editorial Almuzara, 2012. [ Links ]
4 . Cárdenas RVC. Situación del todo de Lidia y de la fiesta en los países de Hispano-América. Editado por García AL. Congreso Mundial Taurino de Veterinaria, Consejo General de Colegios Veterinarios de España, 2017. [ Links ]
5. Crandall KA, Bininda-Emonds OR, Mace GM, Wayne RK. Considering evolutionary processes in conservation biology. Trends Ecol Evol 2000;15:290-295. [ Links ]
6. Cañón J, Tupac-Yupanqui I, Garcia-Atance MA, Cortés O, Garcia D, Fernandez J, Dunner S. Genetic variation within the Lidia bovine breed. Anim Genet 2008;39:439-445. [ Links ]
7. Scherrer HL. Historia del toro bravo mexicano. México, Asociación Nacional de Criadores de Toros de Lidia (ANCTL), 1983. [ Links ]
8. Niño de Rivera L. Sangre de Llaguno, la razón de ser del toro bravo mexicano.1ra ed. México: Punto de Lectura; 2004. [ Links ]
9. Villanueva Lagar JA. San Mateo, encaste con historia. México: Aldus; 2005. [ Links ]
10. ANCTL. Asociación Nacional de Criadores de Toros de Lidia. México. 2019. [ Links ]
11. Censo Agrícola, Ganadero y Forestal 2007. INEGI. http://www.inegi.org.mx [ Links ]
12. Eusebi PG, Cortés O, Dunner S, Cañón J. Genetic diversity of the Mexican Lidia bovine breed and its divergence from the Spanish population. J Anim Breed Genet 2017;134(4):332-339. [ Links ]
13. Eusebi PG, Cortés O, Dunner S, Cañón J. Genomic diversity and population structure of Mexican and Spanish bovine Lidia breed. Anim Genet 2017;8(6):682-685. [ Links ]
14. Eusebi PG, Gardyn OC, Boxberger SD, Ferreras JC. Genetic diversity analysis of the Mexican Lidia bovine breed population and its relation with the Spanish population by using a subset of SNPs under low gametic disequilibrium. Rev Mex Cienc Pecu 2018;9(1):121-134. [ Links ]
15. Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, 2nd ed. NY, USA: Cold Spring Harbor Laboratory Press, Cold Spring Harbor, 1989. [ Links ]
16. Purcell SM, Neale B, Todd-Brown K, Thomas L, Ferreira MAR. et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559-575. [ Links ]
17. Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online 2005:1, 47. [ Links ]
18. Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res 2009;19:55-64. [ Links ]
19. Alexander DH, Lange K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC bioinformatics 2011;12:246. [ Links ]
20. Francis RM. Pophelper: an R package and web app to analyze and visualize population structure. Mol Ecol Resour 2016. [ Links ]
21. Purfield DC, Berry DP, McParland S, Bradley DG. Runs of homozygosity and population history in cattle. BMC Genetics 2012;13:70. [ Links ]
22. Crow JF, Kimura M. An Introduction to Population Genetics Theory. New York. USA: Harper & Row, Publ; 1970. [ Links ]
23. Upadhyay MR, Chen W, Lenstra JA. et al. Genetic origin, admixture and population history of aurochs (Bos primigenius) and primitive European cattle. Heredity 2016;118:169-76. [ Links ]
24. Cortés O, Tupac-Yupanqui I, Dunner S, Fernández J, Cañón J. Y chromosome genetic diversity in the Lidia bovine breed: a highly fragmented population. J Anim Breed Genet 2011:491-498. [ Links ]
25. Wright S. The genetical structure of populations. Ann Eugenics 1951;15:323-354. [ Links ]
Received: February 23, 2019; Accepted: October 07, 2019











 texto em
texto em 


