Introduction
Escherichia coli (E. coli) can colonize the gastrointestinal tract of humans and animals without causing damage, there are pathogenic strains that constitute a heterogeneous group of organisms with different virulence properties, serotypes O:H and epidemiology. Based on their specific virulence factors and phenotypic characteristics, they have been subdivided into six pathogenic groups: enteropathogenic E. coli (EPEC), enteroaggregative E. coli (EAEC), enterotoxigenic E. coli (ETEC), diffuse adhesion E. coli (DAEC), E. coli enteroinvasora (EIEC) and enterohemorrhagic E. coli (EHEC)1; one of the most important virulence factors in E. coli have been the Shiga toxins (Stx) and the products of the pathogenicity island as the site of the disappearance enterocyte (LEE), these bacteria are called E. coli producer of Shiga Toxin (STEC), a common type of STEC is E. coli O157:H7, but there are other serotypes No-O157:H72, these have been detected in different products, such as meat, dairy products, fish, seafood, beverages, ice or legumes3; however, they have been implicated in outbreaks associated, mainly, with the consumption of bovine meat, they are the main reservoir4, it is found with prevalences in healthy cattle between 7-30 %; it seems that these strains are not pathogenic for animals, although some researchers find them more frequently in those tha have diarrhea5, so the meat can be contaminated with fecal matter that contains E. coli by poorly processing the carcass in the slaugtherhouses.
E. coli can exchange genetic material through mobile genetic elements (MGEs) such as plasmids, transposons and integrons, which facilitates their adaptation to new and adverse environments that contributes to intestinal or extraintestinal disease. They differ in their virulence, resistance, incidence and severity; however, they can not only be attributed to these factors, but they are the result of their interaction with the host factors and the environment4.
In the case of antimicrobials, these are used in animals for three main purposes: growth promotion, prophylactic measures and as therapy when disease occurs6,7, which contributes to the survival of resistant strains. New resistance mechanisms are appearing, which are spread around the world and endanger our ability to treat common infectious diseases, adding the consequent increase in the prolongation of the disease, disability and deaths8. Some of these E. coli are present in people, animals and the environment (water, soil and air) and can be transmitted from people to animals and the other way around, including through the consumption of products of animal origin; so an inadequate management in the control of infections, conditions, poor sanitation and inadequate manipulation of food, promote the spread of antimicrobial resistance8.
The impact of the disease caused by STEC has created the need to increase preventive measures of food handling and surveillance of outbreaks9. In addition to traditional epidemiological investigations, the main molecular method for surveillance has been pulsed field electrophoresis (PFGE), which is the method recommended by the Center for Disease Control and Prevention (CDC)10, the use of this technique is crucial in STEC infections11. In Mexico, there are STEC serotypes in cattle that may be involved in Foodborne Diseases12,13, so in this study they were characterized and related from different origins to determine the genetic diversity present in Mexico.
Material and methods
Isolates
A total of 32 isolates of E. coli obtained from cattle carcasses and feces were used in three municipal slaugtherhouses (A, B and C) in central Mexico. These isolates belonged to 16 serotypes, as previously determined with serotyping agglutination assays, using 96-well microtiter plates and rabbit sera (SERUNAM, Mexico City, Mexico) obtained against 187 somatic antigens and 53 flagellar antigens for E. coli and against 45 somatic antigens for Shigella species. All isolates were retrieved from frozen stock cultures and grown in Mac Conkey Agar and incubated at 37 °C for 24 h, a colony with the typical morphology was selected and harvested in trypticase soy broth for further characterization.
Antimicrobial susceptibility testing
The evaluation of bacterial resistance to antimicrobials was carried out using the Kirby-Bauer technique standardized by the Clinical Laboratory Standars Institute (CLSI). The control strain was E. coli ATCC 25922 and was used: amikacin (AK 30 μg), ampicillin (AM 10 μg), carbencillin (CB 100 μg), cefalotin (CF 30 μg), cefotaxime (CTX 30 μg), ceftriaxone (CRO 30 μg), chloramphenicol (CL 30 μg), gentamicin (GE 10 μg), netilmicin (NET 30 μg), nitrofurantoin (NF 300 μg) pefloxacin (PEF 5 μg) and trimetropima-sulfamethoxazole (STX 25 μg). (Sensidisks, Gram Negative BIO-RAD Cat # 71080280). The bacterial inoculum was adjusted to a turbidity equivalent to 0.5 of the McFarland scale and plated in Mueller Hinton Agar with a sterile swab and on the inoculum were placed in the sensidiscs. They were incubated for 24 h at 37 °C. All isolates were classified as resistant, intermediate or susceptible as previously described. Isolates were considered as MDR when resistance to three or more kinds of antimicrobial agents were presented14.
Pulsed fields gel electrophoresis (PFGE)
The clonal relationship of the E. coli isolates was performed using the PFGE technique according to the protocol standardized by the CDC (Center for Diseases and Prevention Atlanta, GA, USA) for the PulseNet Network, as a marker the strain was used of Salmonella Serovar Braenderup H981215. The electrophoresis conditions were those established according to the protocol of the PulseNet network suggested for the Cheef Dr-II model (Bio-Rad, Munich, Germany) and are as follows: Initial time: 2.2 sec, final time: 63.8 sec, Voltage: 6v / cm2, run time: 21 h10. The gel was stained with 200 mL of ethidium bromide for 40 min at 100 rpm and subsequently washed with 200 mL of distilled water for 1 h at 100 rpm.
The band pattern was observed under ultraviolet light (UV) and the digital image of the PFGE patterns was taken using a SmartGeL II photodocument (Sagecreation). The obtained bands were analyzed using the Software BioNumerics Version 7.5 (Applied Maths, Austin, TX), this program allowed to compare the band patterns obtained in different gels and to identify the different restriction profiles. The establishment of the genetic relationship between the strains was carried out by applying the coefficient of similarity of Dice's between the different patterns of bands obtained, and a dendrogram was constructed by means of the UPGMA method (Underweighted Pair-Group Method with Arithmetic Averages) with tolerance values of 1.5 %.
Results
In the antimicrobial resistance 75 % (24 of 32) showed resistance to some antibiotic, of these 28.1 % (9/32) of the isolates was MDR, 84.3 % (27/32) presented intermediate profile in at least one antibiotic and 12.5 % (4/32) was sensitive to all antimicrobials. The resistance frequency of CB is 46.9 % (15/32), CF 50 % (16/32), AK 37.5 % (12/32), GE 28.1 % (9/32), AM 21.9 % (7/32) and CTX 3.1 % (1/32), however several isolates showed intermediate profile as in AM 56.3 % (18/32), PEF 46.9 % (15/32) and only SXT was sensitive in all the isolates (Figures 1 and 2).
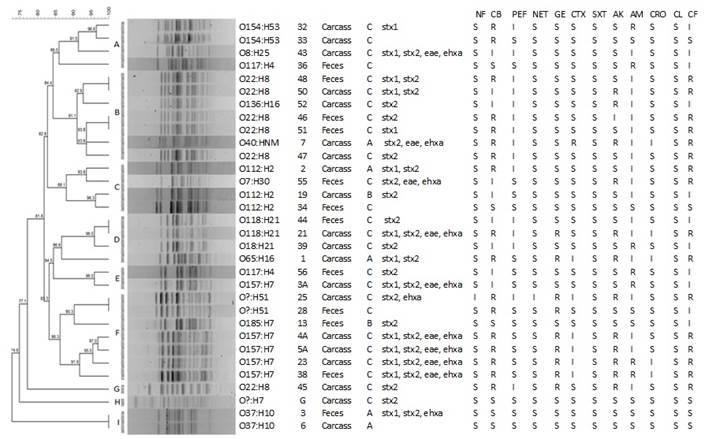
In the columns: the serotype, identification, origin, slaughterhouse, virulence profile and resistance profile (NF-Nitrofurantoin, CB-Carbencillin, PEF- Pefloxacin, NET-Netilmicin, GE-Gentamicin, CTX-Cefotaxime, STX-Trimetropim-sulfamethoxazole, AK-Amikacin, AM-Ampicillin, CRO-Ceftriaxone, CL-Chloramphenicol and CF-Cefalotin).
Figure 1 PFGE profile (XbaI) obtained from STEC isolates from a carcasses and feces of cattle in three municipal slaughterhouses of central of Mexico

Figure 2 Frequency of antimicrobial resistance from STEC isolates from a carcasses and feces of cattle in three municipal slaughterhouses of central of Mexico
The genetic relationship between the different isolates of E. coli was investigated by Genotyping with the XbaI-PFGE enzyme; of the 32 isolations obtained from carcasses and feces in three slaugtherhouses (A, B and C) of bovine in the Center of Mexico, 27 PFGE pulsotypes were observed; 7 clusters were formed with 2 or more isolates (A-F and I) and two integrated with a strain (G and H) these with percentages of similarity higher than 85 % (Figure 2). Clusters D, E, F and G grouped of the MDR isolates. Five clonal pulsotypes were identified, in the clonal pulsotypes 1 and 2 are of serotype O22:H8 (cluster B) the first comes from different sources such as carcasses (isolated 48) and feces (isolated 50); the second are feces (isolated 46 and 51); however, the resistance profile is different; the third clonal pulsotypes (Cluster D) of serotype O118:H21 is from feces and carcasses (isolates 44 and 21) but with different resistance profile; the fourth pulsotypes (Cluster F) is from carcasses and feces (isolates 25 and 28) and belong to the serotype O?:H51 however one of them is MDR; the fifth pulsotypes (Cluster I) is feces and carcasses (isolates 3 and 6) and are of serotype O37:H7, both sensitive to all antimicrobials; the MDR isolates are found in different pulsotypes, which shows their diversity (Figure 2).
Discussion
STEC contributes to 265,000 cases of foodborne illness annually in the United States, the antimicrobial drug resistance among STEC has been reported but is probably underestimated16. The antimicrobial resistance is one of the most serious problems in medical care17, representing more than 700,000 deaths per year; this is because of the acceleration of antimicrobial resistance genes (ARGs) into bacteria via de novo mutation. In addition of the horizontal transfer of the mobile genetic elements (MGEs) as plasmids, transposons and integrons18, which causes an emergency in foodborne bacteria, is a public health issue mainly in meat, as it is considered an important carrier for antimicrobial-resistant E. coli17.
In the present study STEC strain is identified in cattle carcasses and feces, where 28.1 % are MDR, so their presence is a risk. However, all isolates are sensitive to STX but its use as a treatment in human infections can induce the production of the shiga toxin19 leading to more serious disease; several authors indicate that foods of animal origin are often contaminated, and point out the need to improve the meat process and indicate that carcasses contamination occurs due to poor handling of the intestinal content of carrier cattle2.
A study conducted in Mexico in isolated from beef samples found 92.4 % of strains MDR; unlike our study, they found strains that exhibited multidrug resistance patterns from 7 to 9 antibiotics, simultaneously (44 %, n =77)20, this could be because the samples were obtained in supermarkets. However, it has been seen that cross contamination is of the utmost importance, the exposure at the sale stage represents a global combination of the entire production chain (from farm to fork) which represents a greater risk to public health17. In STEC, isolates were outbreak associated from the Michigan Department of Health and Human Services (MDHHS) Reference Laboratory (Lansing, MI, USA), collected during 2010-2014 found resistance to ampicillin, trimethoprim/sulfamethoxazole and ciprofloxacin an indicate that the frequency of resistance vary according to their location and source; although, they do not observe differences in frequency in localities16, similar to what happens in Mexico.
The permanent surveillance of antibiotics used in livestock is of great importance to determine the presence and prevalence of resistant strains in the carcasses, this puts human health at risk8; beef contaminated with antimicrobial-resistant bacteria, when not properly handled and cooked, could transfer their resistance genes to other pathogens in addition to its toxins, which could lead to a difficult disease to treat21. Cooking had the greatest impact on the reduction of the hazards17; however, there are studies in which E. coli survived to heating at 70°C, and also showed that it retains its characteristics and genes that encode the resistance, and it is also possible to transfer by electroporation, so there is a risk of a natural transformation22. Therefore, a strategy to reduce the risk of STEC infections consists of reducing the prevalence in livestock with adequate manufacturing practices as well as improving the handling of slaugtherhouses.
In this study, several STEC elements of importance are observed, however in serotype O157:H7 it can present asymptomatically or it can develop diarrhea, haemorrhagic colitis and/or haemolytic uremic syndrome2 and resistance of 5 antibiotics: AK, AM, CB, CF, and GE, in the serotype O117:H4 were only resistant to AM, this is an emerging serotype that causes sepsis in humans and it has also been found in cattle with diarrhea23 epidemiologically, its presence is important. In serotype O22:H8 presents a resistance to AK, CB, CF and GE and it has been associated with disease in humans and it has been found in a serotype in Brazil24, France25 and Argentina26; in the Valley of Culiacán, northwest of Mexico, of the STEC O157 and non-O157 strains tested that are considered resistant to antimicrobials belonging to classes such as aminoglycosides, beta-lactams and cephalosporins19. In Tamaulipas, Mexico, in retail meat samples, 92.4 % of the strains were resistant to cefalotin, ampicillin, cefotaxime and nitrofurantoin20) with these reports it can be said that in Mexico they are found different but similar resistance profiles in the study area. In another study conducted in Ethiopia, the most frequent resistance isolates were cefoxitin, ampicillin and amoxicillin27. In China from 2000 to 2012 in E. coli isolates from two intensive poultry farms were highly resistant to SXT, AM and GM; they find resistance in sulfonamides but this is because the poultry production units are subject to problems diarrheal of bacterial and parasitic origin; likewise, non-O157 STECs isolated from humans and animals have shown resistance to multiple antimicrobials, including resistance to trimethoprim-sulfamethoxazole28. In Egypt, a study of isolates of animals and humans (diarrheal children) showed resistance to one or more antibiotic agents, and they were resistant to cephalothin regardless of their origin (food or human), find closely related isolates (97 %) in feces of Swiss cattle and human disease in Germany29 so the use of antibiotics in livestock environments can affect and cause the presence of multiresistant bacteria.
The serotype O157:H7 was found only in the slaugtherhouse C, 4 are from carcasses and 1 of feces, this confirms the importance of the handling that there is in the carcasses in slaugtherhouses and the reason why presence in carcasses demonstrates the crossed contamination that exists; also, that these were taken at different times and none of these isolates are clones becasuse they have an 83.3 % similarity. In a study conducted on meat products over the period 2004-2013, they found a similarity of 67 % and found no clonal relationship between the isolates30, confirming that meat products are an important source of contamination due to inadequate management in the process of obtaining products and by-products of bovine origin31; isolates have been found in humans with hemolytic uremic syndrome, sauces and cooked and raw beef which have the same origin and mention that 12% of infections in humans in Argentina are of Bovine origin32.
In another study carried out in 4 rural farms in Culiacán, Mexico, it was found that the O157:H7 serotype shows 6 pulsetypes in one of the identified clones from cattle, sheep and birds12; In another study carried out on a federal inspected slaughterhouses in Mexico in 2009-2010, they found 49 pulsotypes in 97 isolates of O157:H7 grouped into 3 clusters with 80% similarity13, so this study, despite having fewer isolates, shows greater diversity in Mexico; this may be because one of the studies was conducted in rural farms of Culiacán and the other were driven by Federal Inspections on slaugtherhouses, which has more control in the process of obtaining meat. In this study the isolates were from municipal slaugtherhouses, where management is deficient and animals from different production units arrive. They supply one of the main metropolitan areas in Mexico, so the presence of STEC in cattle can be an important factor of contamination in the process of obtaining meat. In serotype O22:H8, the six isolates presented diversity and were grouped into four pulsetypes: the first is carcasse and feces, the resistance profile is different; in the second profile, the two isolates are feces, so probably these isolates belong to animals that are in the same production unit (91.1 % similarity). These data demonstrate the presence of this serotype in slaugtherhouse C, which indicates a risk; this has been associated with severe disease in humans. The presence of 3 different isolates of serotype O112:H2 was found, two of them are from carcasse and one is from feces, but these were presented in the 3 slaugtherhouses (A, B and C) so in the center of Mexico it is present this serotype as well. Each slaugtherhouse has a variant, they have 88.1 % similarity, and this serotype has been found in beef33 and hamburgers34, so its presence in carcasse is to be expected. The clones present of serotype O118:H21 were from carcasse and feces, one of them is MDR; they were taken in the same slaugtherhouse (C), however at different times, so there is the possibility that the animals are of the same production unit or this serotype is not so diverse.
In a study carried out in north-east of Englad, they drove an epidemiological investigation with the infections produced by STEC and associated with it, the consumption of meat products and livestock production units. By sequencing the isolates they found epidemiological links between the clinical cases, the butchers and the farm that supplied the product, which showed the cross-contamination with ground beef and other products35.
Conclusions and implications
This confirms the importance of the management that occurs in livestock production units, the cattle is considered the main reservoir, and the prudent use of antibiotics is recommended to avoid resistance in addition to maintaining the efficacy of the drug; however, if it is used improperly, it could cause potential adverse effects, clinical implications and the acceleration in the development of resistant bacteria that can colonize the human gastrointestinal tract through the food chain, so that serotypes resistant to antibiotics can be found and with the presence of toxins, which would represent a severe public health problem.











 texto em
texto em 


