Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO
Share
Revista mexicana de ciencias pecuarias
On-line version ISSN 2448-6698Print version ISSN 2007-1124
Rev. mex. de cienc. pecuarias vol.11 n.4 Mérida Oct./Dec. 2020 Epub Mar 02, 2021
https://doi.org/10.22319/rmcp.v11i4.5137
Articles
Molecular detection of bovine coronavirus associated with the bovine respiratory complex in beef cattle in the Mexicali Valley, Baja California, Mexico
a Universidad Autónoma de Baja California. Instituto de Investigaciones en Ciencias Veterinarias. Km. 3.5 Carretera a San Felipe, Fraccionamiento Campestre, 21386, Mexicali, Baja California, México.
b Universidad Autónoma de Sinaloa. Facultad de Medicina Veterinaria y Zootecnia, Culiacán Sinaloa, México.
The bovine respiratory complex (BRC) is the leading cause of disease and death in beef cattle worldwide. It is a multifactorial infectious syndrome caused by different viruses and bacteria that reduce the productive efficiency and cause economic losses. In Mexico, BRC has been reported in all regions where cattle are fattened; however, these reports lack information on the presence of bovine respiratory coronavirus (BCV). This makes it necessary to have reliable and accurate diagnostic tools for detecting the presence of BCV in beef cattle fattened in Mexico, in order to propose appropriate sanitary measures for their clinical management. In this work, a real-time-PCR molecular diagnostic platform (rt-PCR) was developed to amplify a fragment of the BCV S protein in nasal exudate samples. When applying the rt-PCR platform for BCV in seemingly healthy beef cattle with signs of respiratory disease associated to BRC, 19/50 (38 %) were found to be positive, confirming the presence of this virus in the cattle of the region. The results of this work constitute the first report on the presence of the BCV associated to the BRC in the cattle region of northwestern Mexico and establish the bases for future research about the role that this virus plays in the presentation of the pathology of the BRC in beef cattle exploitation systems in this region and across the country.
Key words Bovine Respiratory Coronavirus; Bovine Respiratory Complex; PCR; Protein S; Beef cattle
El complejo respiratorio bovino (CRB) es la principal causa de enfermedad y muerte en el ganado de engorda en todo el mundo. Es un síndrome infeccioso multifactorial provocado por distintos virus y bacterias que disminuyen la eficiencia productiva y ocasionan pérdidas económicas. En México, el CRB se ha reportado en todas las regiones donde se engorda ganado; sin embargo, esos reportes carecen de información sobre la presencia del coronavirus respiratorio bovino (CVB), haciendo necesario contar con herramientas de diagnóstico confiables y precisas para detectar la presencia de CVB en el ganado que se engorda en México, para proponer las medidas sanitarias apropiadas para su manejo clínico. En este trabajo, se desarrolló una plataforma de diagnóstico molecular de PCR en tiempo real (rt-PCR) que amplifica un fragmento de la proteína S del CVB en muestras de exudado nasal. Al aplicar la plataforma rt-PCR para CVB en bovinos de engorda en aparente estado de salud y con signos de enfermedad respiratoria asociados a CRB se encontró que 19/50 (38 %) resultaron positivos, confirmando la presencia de ese virus en el ganado de la región. Los resultados de este trabajo significan el primer reporte sobre la presencia del CVB asociado al CRB en la región ganadera del noroeste de México y sienta las bases para futuras investigaciones sobre papel que juega este virus en la presentación de la patología del CRB en los sistemas de explotación de bovinos de engorda en nuestra región y el país.
Palabras clave Coronavirus respiratorio bovino; Complejo respiratorio bovino; PCR; Proteína S; Bovinos de engorda
Introduction
Bovine Respiratory Complex (BRC) is considered to be the main cause of clinical disease and death in feedlot cattle worldwide, and thereby, of economic losses for the farmers1. Traditionally, BRC is associated with single or combined disease of bovine respiratory syncytial virus (RSV), type 1 bovine herpes virus (BHV), bovine parainfluenza virus 3 (BPI3) and bovine viral diarrhea virus (BVDV), which produce a primary infection with mild clinical signs2. The BRC is also associated with the presence of the bacteria Pasteurella multocida, Mannheimia haemolytica, Histophilus somni, and Mycoplasma bovis, which act as opportunistic pathogens during conditions of stress or primary viral infection3,4. Besides the mentioned viruses, the bovine coronavirus (BCV) has also been reported as a viral agent associated to BRC, causing respiratory disease and reduction in weight gain in beef cattle5-9. The infection produced by BVC has been reported all over the world and is considered an endemic disease in dairy and beef cattle farms7. BVC is transmitted mainly through the fecal-oral route, although it has also been shown to be transmitted through the respiratory route, by inhalation of aerosols containing the viral particles. When BCV enters the gastrointestinal tract, it triggers a clinical picture of diarrhea, dehydration, acidosis and hypoglycemia in young animals10. When BVC is admitted through inhalation, it infects the respiratory epithelium of the nasal turbinates, trachea and lungs. Replication leads to the elimination of the virus in nasal secretions, and the disease produces a picture with clinical signs ranging from absent to severe, including depression, fever, conjunctivitis, respiratory distress, and mild to severe cough11.
In Mexico, the BRC has been reported in all regions where cattle are fattened12,13,14. However, these reports lack information regarding the presence of the BVC in beef cattle farms, being of the greatest importance for developing diagnostic tools that allow to confirm the presence of the BVC in the cattle. The diagnosis of BVC is achieved using different serological techniques10,15 or viral isolation from nasal exudate and biopsies from different tissues10,16,17. The application of molecular techniques for the detection of BVC associated with BRC―including conventional PCR (PCR) and real-time (rt-PCR)―has recently been reported6,10,14. Prominent among these are the platforms that amplify the gene which encodes for the BCV S protein, the most important viral structure for the production of neutralizing antibodies7,18. The S-protein gene is highly conserved among the BCV strains and has been widely used as a target gene for molecular tests for diagnosing this disease in livestock and other animal species, including humans6,19,20.
The aim of this work was the development and use of a platform for molecular diagnosis by rt-PCR to detect a fragment of the gene that codes for the BCV S protein in samples of bovine nasal exudate. The results indicate that the rt-PCR system is highly sensitive and specific for detecting the BVC and can be used in beef cattle exploitation systems in the region and across the country.
Material and methods
This study was conducted at the Diagnostic Laboratories Unit (ULADI) of the Institute of Research in Veterinary Sciences of the Autonomous University of Baja California, Mexicali Campus.
Nasal exudate samples
Fifty (50) nasal exudate samples were collected from beef cattle in stables, belonging to a technified bovine exploitation system located in the Mexicali Valley, Baja California. The samples were obtained from newly admitted animals aged 18 months on average, after less than 30 d of their arrival at the farm. Thirty samples were collected from animals with nasal discharge, cough, depression, or body temperature greater than 38.5°C (Group 1), which were classified as sick, and 20 samples, from apparently healthy animals that showed none of the above21. The nasal exudate samples were collected by deep intranasal route, using broom-type Dacron swabs. Once the samples were taken, each swab was immersed in a tube containing sterile phosphate-base saline (PBS), pH 7.4, and the handle was cut so that the tube could be closed to protect the sample from possible contamination; the corresponding group was identified with a progressive number. Once collected, the samples were transported to the laboratory for processing.
Removal of RNA from nasal exudate
For RNA extraction, Aurum Total RNA Fibrous Tissue Reagent Kits (Bio Rad, Hercules, California, USA) were used according to the manufacturer's instructions. RNA was recovered from each extraction in a volume of 50 µL of the elution solution provided in the reagent kits. The extracted RNA was stored frozen at -20 °C until the time of the rt-PCR.
Oligonucleotide design for BCV
Oligonucleotides were designed from the sequence of the gene that encodes for the S protein of the bovine coronavirus strain R-AH187 (BCV-S), with access number GenBank EF424620.1. The gene corresponds to a 4,090 base pair molecule published in July 2016. The Primer3Plus oligonucleotide design software, version 2006-2007, was used; it is available at: http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi. From the sequence of the BCV-S gene, the positive band oligonucleotide called BCVf with 5'-CTACTTGGAATAGGAGATTG-3' sequence was generated, while for the negative band oligonucleotide called BCVr, the selected sequence was 3'-TACACGGAGAAATTGG-5'; the amplification of these oligonucleotides by rt-PCR generates a product of 132 base pairs and a 36 % GC content, with a dissociation temperature (Tm) of 77.0 ºC for this PCR product. Table 1 shows the characteristics of oligonucleotides. Oligonucleotides were synthesized by GenScript LTD (Piscataway, New Jersey, USA) and were reconstituted with molecular biology grade water equivalent to 10 times the nano molar (nM) concentration value referred to by the manufacturer, to obtain a standard concentration of 100 micro molar (µM). For the rt-PCR tests, the working concentration of the oligonucleotides was set at 10 µM.
Table 1 Sequences and properties of oligonucleotides designed from the BCV-S gene with the reference GenBank EF424620.1
| Oligonucleotide: | BCVf | ||
| Sequence: | CTACTTGGAATAGGAGATTTG | ||
| Start: nucleotide 1337 | Length: 21 pb | Tm: 55.4 ºC | GC: 38% |
| Oligonucleotide: | BCVr | ||
| Sequence: | TACACGGACAGAAATTTGTG | ||
| End: nucleotide 1469 | Length: 20 pb | Tm: 54.3 ºC | GC: 40% |
| Product: | 132 pb | Tm: 77.0 ºC | |
Master mix
In this work it was used the One-Step RT-PCR Script masterbatch I (Bio Rad, Hercules, California, USA) formulated with SYBR Green I fluorophore in a masterbatch solution using both oligonucleotides at a concentration of 400 nM, 2 µl RNA quench and molecular biology grade water for a total reaction volume optimized to 10µL.
Positive RNA controls for BCV (o)
As positive RNA control extracted from the liquid fraction of the Scourgard 4 K7C vaccine (Zoetis, New Jersey, USA), which contains inactivated Hansen strain bovine coronavirus and inactivated G6 and G10 bovine rotavirus strains, enterotoxigenic E. coli K 99 and Clostridium perfringens toxoid type C, was utilized. The procedure for RNA extraction was performed according to the protocol of the Bio Rad Aurum Total RNA Fibrous Tissue set of reagents, using 300 µl of the vaccine. The extracted RNA was divided into 10 µl aliquots and stored in a freezer at -20° C until the RT-PCR tests were performed.
RT-PCR test protocols
The rt-PCR tests were performed on a Bio Rad CFX96 thermal cycler. Denaturation, hybridization and extension parameters were calculated using the Protocol Autowriter tool of the CFX96 package integrated to the thermal cycler, taking into consideration the size of the PCR product, the oligonucleotide sequence and the type of enzyme in the master mix, resulting in an initial step at 50 °C during 10 min for reverse transcription, followed by a denaturation cycle of 95 °C during 3 min, and continuing with 39 denaturation cycles at 95 °C during 10 sec, 20 sec at 51. 0 °C for oligonucleotide hybridization, and 15 sec at 72 °C for extension. Also, for each run, dissociation curve analysis from 65 °C to 95 °C was performed to identify amplification curves within the estimated temperature of 77.0 °C +/- 1 °C of the 132 bp PCR product and discriminate between artifacts other than the amplification of the expected RNA template.
Interpretation of results
rt-PCR test results for BCV were considered positive when the corresponding sample obtained a fluorescent amplification signal before cycle 40, above the threshold control line automatically established by the CFX96 program, and amounting to 10 times the standard deviation of the average fluorescence index generated by all the samples during the first 10 cycles of each run. Results were considered negative when the corresponding sample failed to develop a fluorescent amplification signal above the threshold line of the negative reference control within a maximum of 40 cycles.
Results
RT-PCR standardization for BCV (o)
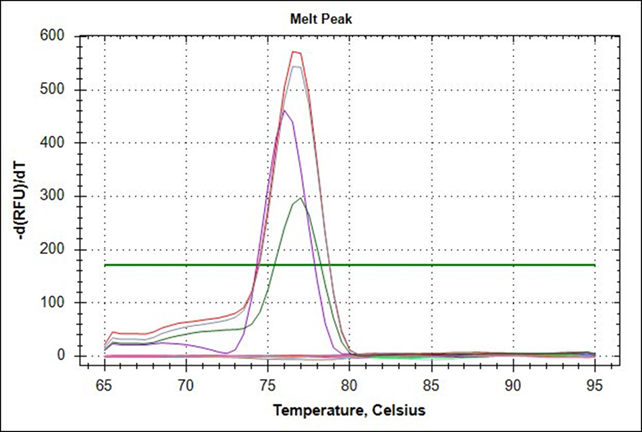
The amplification graph and dissociation curve calculated with Bio-Rad’s CFX96 package for the BVC rt-PCR system showed that the optimal combination of reagents for maximum amplification of the oligonucleotides BCVf and BCVr is achieved at a concentration of 400 nM with 2 µL RNA template. Under these conditions the positive controls extracted from the Scourguard 4 K/C vaccine developed a signal above the threshold line with an average amplification cycle (Cq) of 31.09 in 40 total cycles of each amplification run; the negative controls showed no evidence of amplification (Figure 1). Also, analysis of the dissociation curve (Tm) for the positive control RNA showed a dissociation temperature range between 76.0 and 78.0 °C, with an average temperature of 77.0 °C (Figure 2); these parameters allow considering the rt-PCR test for CVB as valid.
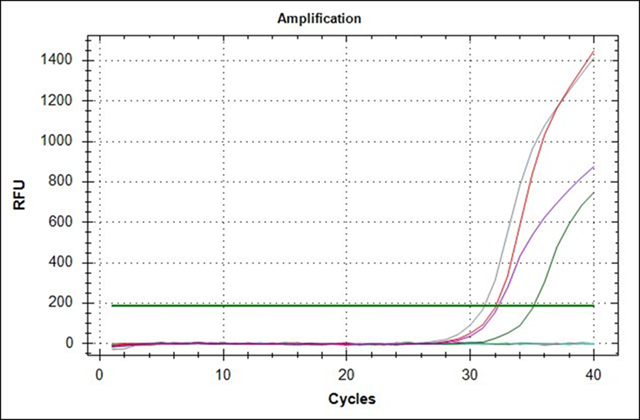
Figure 1 Amplification curve of bovine coronavirus controls extracted from Scourguard 4 K/C vaccine using the oligonucleotides BCVf and BCVr at a concentration of 400 nM with a 2 µl RNA template
RT-PCR results for nasal exudate samples
Fifty RNA samples of nasal exudate from stabled cattle were tested in duplicate, of which 19 (38.0 %) achieved amplification above the threshold line established by CFX96 and were therefore considered positive. Of the samples that were positive, 5 (10.0 %) belonged to Group 1, which corresponds to animals with signs and symptoms associated with the BRC, and 14 (28.0 %), to Group 2, which is made up of animals without signs and symptoms of respiratory disease. The average Cq of the samples of both groups was 34.60 cycles with a range of Cq between 30.87 and 35.95 cycles, and average Tm of 77.0 °C (Figure 2).
Discussion
BCV is a globally distributed pathogenic virus that causes enteric diseases in young calves and winter dysentery in adult cattle. BVC is also implicated in BRC-associated infections in beef cattle. Although infections by BCV produce a mortality smaller than 2 %, the morbidity of this virus can reach 100 % of the animals of a farm, causing respiratory or digestive syndromes that negatively affect the rate of gain of weight or milk production and increase the costs due to veterinary services, antibiotics and other medicines that altogether cause economic losses for the cattle sector10,15.
The development and implementation of the rt-PCR platform for BCV presented here, arises in response to the need for reliable, accurate and rapid diagnostic tools to detect a disease of viral origin that has been reported as part of the BRC; however, due to the large number of signs and symptoms that common pathogens produce, it is difficult to establish precisely the main causal agent of the pathology in an animal or herd22,23; especially when the infectious process develops with minimal or imperceptible symptoms, causing a delay in the initiation of the corresponding therapy, extending the time required to recover the state of health and, therefore, negatively affecting the levels of productivity of the sick animals20,24.
In this work, an rt-PCR platform for detecting and amplifying a fragment of the gene that encodes for the BVC S protein was designed, developed and instrumented, which turned out to be a highly sensitive and specific molecular diagnostic platform for the detection of BCV from nasal exudate samples. Although in this work the viral particles were not quantified, the sensitivity of rt-PCR platforms to BCV from nasal exudate samples has been previously reported as having detection ranges of 102 cDNA25 to 103 cDNA26 copies per reaction, with amplification curves developed after cycle 34 for both studies. This agrees with the average Cq of 34. 60 developed by the samples analyzed in this work; therefore, can be propose that the rt-PCR platform for CVB reported here has an estimated sensitivity between 102 and 103 cDNA copies per reaction.
Due to the genetic properties of the S protein, characterized by a high homology between the different viral strains and a high immunological reactivity22,24, different fragments of the BCV S protein gene have been used as a reference methodological basis for the development of molecular and serological diagnostic platforms for the rapid detection and diagnosis of this virus with high levels of sensitivity and specificity, even in samples containing small amounts of virus, where conventional diagnostic tests may be inconclusive23.
The results obtained indicate that 19 samples (38 %) from both study groups tested positive with the rt-PCR platform for BCV. Notably, while five of the samples from the group of sick animals (n= 30) were positive, 14 of the samples from the group of apparently healthy animals (n= 20) were positive to the tests. Contrary to what was anticipated, 70 % of the samples from the group of apparently healthy animals were positive to the rt-PCR platform for BCV. This may be because BCV can infect up to 45 % of newly arrived cattle without showing obvious signs or symptoms of disease27,28. However, apparently healthy cattle with nasal shedding of BCV have been shown to be 1.6 times more likely to suffer at least one episode of respiratory disease and 2.2 times more likely to develop lung lesions than animals that do not shed virus by this route7,29; therefore, these animals may have been incubating the virus while not yet developing the clinical respiratory profile characteristic of BRC.
The prevalence of 38 % is similar to that reported in other regions of the world. In a study carried out in Australia in beef cattle for export, nasal exudate samples were analyzed using an rt-PCR platform similar to the one used here, finding a prevalence of 40.1 % for BVC, followed by 0.4 % for BVDV, 0.3 % for IBR, 0.3 % for RSV and 0.3 % for BPI3, evidencing the magnitude of the influence of BCV on the occurrence of respiratory disease of BRC in that country29. Likewise, these results show a higher positive rate than the one reported in Ireland, where a study was carried out to establish the prevalence of pathogens associated to BRC from samples of nasal exudate using rt-PCR and found a positive rate of 22.9 % for BCV, 11.6 % for BRSV, 7.0 % for BPI3, 6.1 % for IBR, and 5 % for BVDV, highlighting the fact that BCV is the virus associated to BRC that is most frequently diagnosed in the beef cattle of that country30. BCV is also the most prevalent virus associated with BRC in beef cattle in the United States of America. Prevalence reports of pathogens associated to BRC in nasal exudate samples from beef cattle analyzed with rt-PCR techniques indicate a prevalence of 62.8 % for BCV, followed by BVDV, with 15.7 %; IBR, with 14.9 %; BRSV, with 9.1 %, and BPI3, with 8.3 %23. The prevalence rate in the USA exceeds that reported for Australia and Ireland, as well as that reported for Mexico through this work, mainly because beef cattle farms in the USA house hundreds of thousands of cattle in a given region, where close contact between healthy and sick animals can favor the transmission and persistence of BCV among cattle31.
The results presented here constitute the first report on the presence of the BCV associated to the BRC in the cattle region of northwestern Mexico and place the BCV in the second position of the table of prevalence of virus associated to the BRC previously detected in the area, where the bovine respiratory syncytial virus (BRSV) occupied the first position with 80.6 % of prevalence, followed by the parainfluenza virus 3 (BPI3), with 23.8 %; the bovine herpes virus 1 (BHV)-1, with 20.4 %, and the bovine viral diarrhea virus (BVDV), with 11.3 %15.
Conclusions and implications
It concludes that the BCV is present in the cattle stables of the Mexicali Valley, Baja California. The rt-PCR platform for BCV reported here is a fast, sensitive and specific molecular diagnostic tool to detect BCV in nasal exudate samples from cattle in feedlots. The prevalence of 38.0 % for BCV reported in this work should be the starting point for future researches on the role that this virus plays in the presentation of the BRC pathology in the beef cattle exploitation systems in our region and across the country.
The authors of this paper declare that they have no conflict of interests of any kind.
Literatura citada
1. Hay KE, Morton JM, Schibrowski ML, Clements ACA, Mahony TJ, Barnes TS. Associations between prior management of cattle and risk of bovine respiratory disease in feedlot cattle. Prev Vet Med 2016;(127):37-43. [ Links ]
2. Gershwin LJ, Van Eenennaam AL, Anderson ML, McEligot HA, Shao MX, Toaff-Rosenstein R, et al. Single pathogen challenge with agents of the bovine respiratory disease complex. PLOS ONE 2015;10(11):e0142479. [ Links ]
3. Hilton WM. BRD in 2014: where have we been, where are we now, and where do we want to go? Anim Health Res Rev 2014;(15):120-122. [ Links ]
4. Xue W, Ellis J, Mattick D, Smith L, Brady R, Trigo E. Immunogenicity of a modified-live virus vaccine against bovine viral diarrhea virus types 1 and 2, infectious bovine rhinotracheitis virus, bovine parainfluenza-3 virus, and bovine respiratory syncytial virus when administered intranasally in young calves. Vaccine 2010;(28):3784-3792. [ Links ]
5. Thomas CJ, Hoet AE, Sreevatsan S, Wittum TE, Briggs RE, Duff GC, Saif LJ. Transmission of bovine coronavirus and serologic responses in feedlot calves under field conditions. Am J Vet Res 2006;(67):1412-1420. [ Links ]
6. Bidokhti MR, Travén M, Ohlson A, Baule C, Hakhverdyan M, Belák S, et al. Tracing the transmission of bovine coronavirus infections in cattle herds based on S gene diversity. Vet J 2012;(193):386-390. [ Links ]
7. Saif LJ. Bovine respiratory coronavirus. Vet Clin North Am Food Anim Pract 2010;(26): 349-364. [ Links ]
8. Hick PM, Read AJ, Lugton I, Busfield F, Dawood KE, Gabor L, et al. Coronavirus infection in intensively managed cattle with respiratory disease. Aust Vet J 2012;(90):381-386. [ Links ]
9. Liu L, Hagglund S, Hakhverdyan M, Alenius S, Larsen LE, Belák S. Molecular epidemiology of bovine coronavirus on the basis of comparative analyses of the S gene. J Clin Microbiol 2006;(44):957-960. [ Links ]
10. Oma VS, Travén M, Alenius S, Myrmel M, Stokstad M. Bovine coronavirus in naturally and experimentally exposed calves; viral shedding and the potential for transmission. Virol J 2016;(13):1-11. [ Links ]
11. Park SJ, Kim GY, Choy HE, Hong YJ, Saif LJ, Jeong JH, et al. Dual enteric and respiratory tropisms of winter dysentery bovine coronavirus in calves. Arch Virol 2007;(152):1885-1900. [ Links ]
12. Figueroa-Chávez D, Segura-Correa JC, García-Márquez LJ, Pescador-Rubio A, Valdivia-Flores AG. Detection of antibodies and risk factors for infection with bovine respiratory syncytial virus and parainfluenza virus 3 in dual-purpose farms in Colima, Mexico. Trop Anim Health Prod 2012;(44):1417-1421. [ Links ]
13. Solís-Calderón JJ, Segura-Correa JC, Aguilar-Romero F, Segura-Correa VM. Detection of antibodies and risk factors for infection with bovine respiratory syncytial virus and parainfluenza virus-3 in beef cattle of Yucatan, Mexico. Prev Vet Med 2007;(82):102-110. [ Links ]
14. Rodríguez-Castillo JL, Lopez-Valencia, G, Monge-Navarro FJ, Medina-Basulto GE, Hori-Oshima S, Cueto-González SA, et al. Detection and economic impact related to bovine respiratory disease, shrink, and traveling distance in feedlot cattle in Northwest Mexico. Turk J Vet Anim Sci 2017;(41):294-301. [ Links ]
15. Amer HM, Wahed AAE, Shalaby MA, Almajhdi FN, Hufert FT, Weidmann M. A new approach for diagnosis of bovine coronavirus using a reverse transcription recombinase polymerase amplification assay. J Virol Methods 2013;(193):337-340. [ Links ]
16. Decaro N, Mari V, Desario C, Campolo M, Elia G, Martella V, et al. Severe outbreak of bovine coronavirus infection in dairy cattle during the warmer season. Vet Microbiol 2008;(126):30-39. [ Links ]
17. Toftaker I, Toft N, Stokstad M, Solverod L, Harkiss G, Watt N, et al. Evaluation of a multiplex immunoassay for bovine respiratory syncytial virus and bovine coronavirus antibodies in bulk tank milk against two indirect ELISAs using latent class analysis. Prev Vet Med 2018;(154):1-8. [ Links ]
18. Workman AM, Kuehn LA, McDaneld TG, Clawson ML, Chitko-Mckown CG, Loy JD. Evaluation of the effect of serum antibody abundance against bovine coronavirus on bovine coronavirus shedding and risk of respiratory tract disease in beef calves from birth through the first five weeks in a feedlot. Am J Vet Res 2017;(78):1065-1076. [ Links ]
19. Singasa K, Songserm T, Lertwatcharasarakul P, Arunvipas P. Molecular and phylogenetic characterization of bovine coronavirus virus isolated from dairy cattle in Central Region, Thailand. Trop Anim Health Prod 2017;(49):1523-1529. [ Links ]
20. Bok M, Miño S, Rodriguez D, Badaracco A, Nuñes I, Souza SP, et al. Molecular and antigenic characterization of bovine Coronavirus circulating in Argentinean cattle during 1994-2010. Vet Microbiol 2015;(181):221-229. [ Links ]
21. Villagómez-Cortés JA, Martínez-Herrera DI. Epidemiological evaluation of clinical bovine respiratory disease complex in a tropical Mexican feedlot. ROAVS 2013;(3):315-321. [ Links ]
22. Belouzard S, Millet JK, Licitra BN, Whittaker GR. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 2012;(4):1011-1033. [ Links ]
23. Fulton RW, d´Offay JM, Landis C, Miles DG, Smith RA, Saliki JT, et al. Detection and characterization of viruses as field and vaccine strains in feedlot cattle with bovine respiratory disease. Vaccine 2016;(34):3478-3492. [ Links ]
24. Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol 2016;(3):237-261. [ Links ]
25. Escutenaire S, Mohamed N, Isaksson M, Thorén P, Klingeborn B, Belák S, et al. SYBR green real-time reverse transcription-polymerase chain reaction assay for the generic detection of coronaviruses. Arch Virol (2007);152(1):41-58. [ Links ]
26. Amer HM, Almajhdi FN. Development of a SYBR Green I based real-time RT-PCR assay for detection and quantification of bovine coronavirus. Mol Cell Probes (2011);25(2):101-107. [ Links ]
27. Hick PM, Read AJ, Lugton I, Busfield F, Dawood KE, Gabor L, et al. Coronavirus infection in intensively managed cattle with respiratory disease. Aust Vet J 2012;(90):381-386. [ Links ]
28. Lathrop SL, Wittum TE, BrocK KV, Loerch SC, Perino LJ, Bingham HR, et al. Association between infection of the respiratory tract attributable to bovine coronavirus and health and growth performance of cattle in feedlots. Am J Vet Res 2000;(61):1062-1066. [ Links ]
29. Moore SJ, O'dea MA, Perkins N, O'hara AJ. Estimation of nasal shedding and seroprevalence of organisms known to be associated with bovine respiratory disease in Australian live export cattle. J Vet Diagn Invest 2014;(27):6-17. [ Links ]
30. O'Neill R, Mooney J, Connaghan E, Furphy C, Graham DA. Patterns of detection of respiratory viruses in nasal swabs from calves in Ireland: a retrospective study. Vet Rec 2015;175(14):351. [ Links ]
31. Wolfger B, Timsit E, White BJ, Orsel K. A systematic review of bovine respiratory disease diagnosis focused on diagnostic confirmation, early detection, and prediction of unfavorable outcomes in feedlot cattle. Vet Clin North Am Food Anim Pract 2015;(31):351-65. [ Links ]
Received: October 30, 2018; Accepted: September 09, 2019











 text in
text in