The identification of genes encoding complex traits has traditionally been achieved by genome-wide scanning and the candidate gene approach, but these methods do not constitute a reliable strategy for the systematic exploration of a genetic network that causes phenotypic variation in complex traits1. Protein networks provide a systems-level overview of genetic organization and enable the functional modules underlying complex traits to be dissected, which facilitates the prediction of novel candidate genes for a trait2. In cattle, some approaches related to interaction networks have been utilized to identify candidate genes related to phenotypic differences such as marbling3, genes involved in estrus (behavior) in dairy cattle4, and single nucleotide polymorphisms (SNPs) associated with growth traits in Mexican Charolais cows5.
Reproduction is an essential element of livestock production, and fertility traits are of particularly significant economic importance; it is a very complex process that involves numerous consecutive events, including gametogenesis, fertilization, and early embryo development, that must be accomplished in a well-orchestrated manner to achieve a successful pregnancy6.
An improved understanding of the mechanisms that control fertility traits at the organ, cellular, and molecular levels could aid the development of strategies to improve and/or monitor fertility4. The objective of this work was to conduct a search guided by a functional interaction network to identify key genes controlling reproductive traits in cattle and explore genetic variation in those identified genes with potential to be associated with reproductive traits.
A literature review was conducted, and the Genie software (http://cbdm-01.zdv.uni-mainz.de/~jfontain/cms/?page_id=6) was used to perform PubMed-based text mining of genes that had been previously associated with bovine reproductive traits (reference genes). To identify and prioritize candidate genes for the functional network, the interactions of the reference genes were extracted, and the degree of association with reproduction (DAR) was calculated for each of the genes in the subnet as follows:
Where Wij is the weight of the linkage connecting protein i and reference protein j, and Pij is the number of links connecting protein i and reference protein j (excluding itself). Thus, the probability that each of these proteins is associated with reproduction was evaluated based on their interactions with genes whose biological function had already been associated with this trait5. To select the candidate genes associated with phenotypic variations in reproductive traits, the DAR score was used to calculate the positive predictive value (PPV), which indicates the likelihood that a gene is associated with reproduction, so the selection criterion was the highest value of PPV obtained in this analysis, i.e., 0.35,7.
From candidate genes, the Ubiquitin B (Ubb) gene was selected as a target. The genetic variation in the Ubb gene was investigated using eleven DNA samples from four different cattle breeds (3 Holstein, 2 Charolais, 3 Brahman and 3 Angus). Primers UBB-F 5’-GAGAGATTTGTGAGAGATCTTGACG-3’ and UBB-R 5’-CCATTTTAACCTGTTGAGTACCCA-3’ were designed to cover and resequence the bovine Ubb gene (GenBank accession number AC_000176.1). The resulting PCR fragments were purified using Exo-SAP-it (Thermo Fisher Scientific, Waltham, MA, USA), and bidirectional sequencing was achieved using the BigDye® Terminator procedure and an ABI PRISM 3100 Genetic Analyzer DNA sequencer (Applied Biosystems, Foster City, CA, USA). Sequences were aligned with ClustalX 2.0.88.
The presence of SNPs in the resulting sequences was determined by visual inspection of the sequence chromatograms, and the SNPs were defined according to their presence in the screening population associated with the three expected genotypes.
Amplification-created restriction sites coupled to PCR (PCR-ACRS) were designed to genotype the nonsynonymous SNP rs110366695 identified in the previous sequencing screening, and following PCR, the fragments were digested using 2.5 U of Hinf I enzyme and analyzed on a 2.5% agarose gel. The following digestion patterns were observed: 210+132+130+18 bp (allele G) and 210+155+150 bp (allele T).
A population of sixty-seven young Angus and Charolais bulls were genotyped with PCR-ACRS. The allelic and genotypic frequencies were calculated for each breed, and deviations from Hardy-Weinberg equilibrium were tested by running GENEPOP statistical package version 4.29.
A set of 385 reference genes associated with reproductive traits in cattle, through SNPs, expression profiles or their biological function, were identified. According to the PPV, the genes that presented a DAR≥11 had a greater than 33% probability of being associated with reproductive traits in cattle, and those meeting this criterion belonged to the ubiquitin family: Ubiquitin C (Ubc; Gene ID: 444874) and B (Ubb; Gene ID: 281370).
The importance of these proteins in the topology of the interaction network was determined according to the number of interactions; in this case, Ubb and Ubc have 3,775 interactions of 3,856 possible, so they are very important hubs. According to BiNGO, a Biological Network Gene Ontology tool (https://www.psb.ugent.be/cbd/papers/BiNGO), in the subnet that forms Ubb and Ubc, the annotation of Gene Ontology 51094, "positive regulation of developmental process", is overrepresented with a p-value of 4.8 E-09. This result makes sense and relates to reproduction in cattle, as this term of biological process refers to any process that activates or increases the rate or extent of development and whose specific outcome is the progression of an organism over time from an initial condition (e.g., a zygote, or a young adult) to a later condition (e.g., a multicellular animal or an aged adult). Figure 1 depicts the Ubb and Ubc interaction with 23 reference gene modules, i.e., genes previously associated with reproduction in cattle8-33.
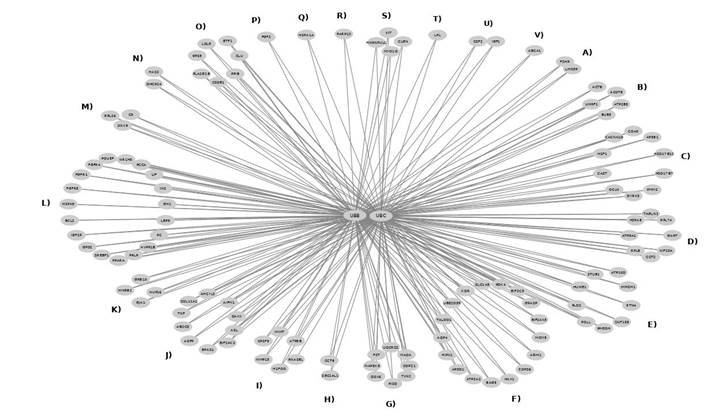
A) Heifer conception rate, SNPs related to interval to insemination. B) Effects of the Well of the Well (WOW) system and embryo density on developmental rates, genes differentially regulated in embryos cultured in vitro. C) Daughter pregnancy rate, heifer conception rate, cow conception rate. D) Term survival in embryos of differentially regulated genes, potential of pretransfer endometrial and embryo gene expression patterns E) Blocking apoptosis in bovine embryos, gene differentially regulated embryos treated with CSF2. F) Immune function and developmental genes expressed in the endometrium, endometrial genes differentially regulated in lactating cows. G) Genes differentially regulated in oocytes compared to 8-cell embryos, global activation of the embryonic genome. H) Cow conception rate. I) Igf1 acts in thermoprotection on bovine embryos, genes differentially regulated in embryos treated with Igf1. J) Endometrial genes differentially regulated in pregnant cows and associations with fertility of lactating dairy cows. K) Genes differentially regulated in oocytes compared to blastocysts, candidate genes for the characterization of the development. L) Liver genes differentially regulated during the transition period, determination of hepatic adaptations occurring from late pregnancy. M) Genes differentially regulated in the oviduct of cows at diestrus compared to estrus. N) Genes in cumulus cells regulated by the LH surge, cumulus cells in development and fertility of oocytes. O) Genes differentially regulated at different stages of oocyte maturation. P) Estimated relative conception rate, net merit, and fat yield. Q) Calving rate (beef cattle), net merit, fat percent, and productive life. R) SNPs related to interval to insemination. S) Embryo development on the blastocyst stage. T) Differential regulation in cumulus cells from in vivo embryos compared to in vitro embryos. U) Antiapoptotic in embryos improves embryo competence. V) Mammary genes differentially regulated during lactation.
Figure 1 Ubb and Ubc interaction network
The ubiquitin protein (Ub) is widespread in all eukaryotic cells, it has a conserved structure which has been interpreted as an indication of its important role in cell metabolism. Through the process of ubiquitination, Ub lead protein degradation and regulate a different biological events including cell cycle progression, membrane-receptor endocytosis, antigen occurrence in the immune system, and even retroviral infection34. Ubiquitination is achieved through the covalent binding of 76-AA, 8.5 kDa ubiquitin to the ε-amino group on the Lys-residues of the substrate via the C-terminal AA residuum of ubiquitin (G76). This process requires ATP hydrolysis and a set of ubiquitin-conjugating factors including ubiquitin-activating (UBA) and conjugating (Ubc) enzymes11.
Among multiple functions of Ub system, those involved in developmental and reproductive processes are relevant. In the former case, there are studies in different models including developmental transitions in Dictyostelium discoideum and development specificity in C. elegans35,36. In chicks it has been involved in embryogenesis, and also in the human myogenesis and brain development34. At reproductive processes level, human Ub has been reported as the main protein in seminal plasma and the ubiquitination system has been implicated with fertility problems in humans and other species including cattle37,38. It has been reported that a high proportion of ubiquitinated spermatozoa in the ejaculates of different species is related to infertility38.
In cattle, increased ubiquitin levels have been associated with both increased levels of damage to sperm DNA and with reduced fertility39. A negative correlation between sperm ubiquitin and sperm count, swirl and % normal morphology has been reported in bulls and evidence that increased ubiquitin levels in bull sperm are predictive of both poor semen quality and fertility has been also found 39. This evidence has allowed the use of ubiquitinated spermatozoa as an useful tool to identify fertility problem40,41.
Even though there is some evidence that the biological mechanisms through ubiquitination systems affect the different species fertility, the process of spermatozoon ubiquitin tagging and the role of this process in sperm biology remains unclarified.
Searching for additional evidence to consider these genes as candidate genes, molecular characterization was achieved on the Ubb gene. According to the NCBI database, the length of the gene is 1898 bp, and it contains one exon at position 841 to 1758. In this database, 19 and 15 SNPs have been reported in the coding and noncoding sequences, respectively, and the amplified 1328-bp fragment enabled the identification of 5 SNPs in the study population, 3 (rs109592218, rs110007734 and rs110366695) in the coding region and 2 (rs720990890 and rs439271103) in the noncoding region. The transversion rs110366695 (G/T) located at exon 1 is particularly interesting because it causes a nonsynonymous functional change, and the GAG codon that produces glutamic acid (Glu, E) changes to UAG, which is a stop codon, thus predicting a truncated protein that is 287 amino acids shorter than the nonmutated protein. Figure 1 shows the allelic frequencies of SNP rs110366695 evaluated in the Angus and Charolais beef cattle breeds; allele G showed the highest frequencies (0.542 and 0.750, respectively). Significant departures (P<0.001) from Hardy-Weinberg equilibrium were identified for the Angus breed, and lower-than-expected numbers of heterozygotes were found for these loci.
To the current knowledge, there have been no previous molecular studies aimed at evaluating the effects of Ubb genetic variation on semen quality despite the demonstrated physiological importance of the Ubb gene. The obtained results support the Ubb gene as a strong candidate gene with genetic variations to be tested for association with reproductive traits. Unfortunately, in Mexico, phenotyping for reproductive traits is not a common practice, and additional efforts must be made to prepare a wide database that allows confirmation of its genetic influence on these traits, particularly of the transversion rs110366695 (G/T).
An analysis-based protein-protein interaction network has been previously validated as a useful tool for identifying causal genes associated with economic traits in bovines and other species. The obtained results provide information about the potential of Ubb and Ubc as candidate genes for reproductive traits, particularly semen quality, and justify further research aimed at exploring both the effects of protein truncation and its potential as a molecular marker.











 texto en
texto en 


