Introduction
Bovine anaplasmosis causes annual economic losses estimated in up to be over $300 million per year, whereas in Latin America those losses were calculated to be approximately $800 million1,2. The causal agent is the intraerythrocytic rickettsia Anaplasma marginale3 and is biologically transmitted by Rhipicephalus and Dermacentor ticks4,5. To date, there are no commercial vaccines available. Wide antigenic diversity and genetic variability are the two major obstacles in the design of immunogens6. Major surface membrane proteins (MSP’s) induced immunity is curtailed by wide genetic diversity and variability of the rickettsia7-10. Type four secretion system (T4SS), VirB7, VirB9, VirB10, ViB11 and VirD4 proteins, together with some outer membrane proteins (OMP’s) of A. marginale have been evaluated for their immunogenic potential11 and have been considered better alternatives to MSP’s candidates.
VirB11 shows an ATPase-like enzymatic activity in Agrobacterium tumefaciens and maintains an ATP-binding Walker-A domain, essential for its ATPase activity12. Immunization with cross-linked A. marginale membrane fragments induced an antibody response against several T4SS proteins including VirB11; this immunization also induced a T cell response against the membrane fragments and their respective recombinant proteins13. It is not yet known however if antibodies generated by natural or experimental infection recognize these proteins, or whether the protein is exposed on the outer surface of the membrane of A. marginale.
Studies of antigenic presentation and immune response have generated deep interest in antigenic peptide-based tools as a newer strategy for inducing immunity. In this sense, multiple antigenic peptides (MAP’s) are highlighted as they allow the combination of epitopes of interest in the same molecular construction, which favors an increase in the sensitivity and specificity of the immune response14.
Based on the importance attributed to VirB11, this work was focused in verifying if the gene is conserved in Mexican strains of A. marginale, its identification and location on the outer surface of initial bodies and testing if antibodies induced by natural or experimental infection react with a multiple antigenic peptide containing one B-cell epitope.
Material and methods
Infected cattle
Five 12-mo old Bos taurus-cross steers were purchased from a local breeder in the Municipality of Cuauhtémoc, west-central Chihuahua State, Mexico which is classified as tick-free by the Servicio Nacional de Sanidad, Inocuidad y Calidad Agroalimentaria (http://www.gob.mx/senasica/documentos/34495). These steers were certified free of tuberculosis and brucellosis by federally certified laboratories. The animals were kept tick-free, and were free of A. marginale as certified by endpoint nPCR for msp5 gene.
The CENID-PAVET laboratories and stalls are located on the outskirts, within the city limits of the small urban area of Progreso, in the Jiutepec Municipality of the state of Morelos, central Mexico. The quarters are free of ticks. Tick treatment is required for animals entering these quarters and all animals housed in outdoor stalls are periodically sprayed for fly control. All animals used in these experiments were housed at the Cattle Isolation Unit of the CENID-PAVET, a tick and fly-proof confinement stall, which includes a double door, screened windows and an electronic indoor insect killer.
Steers 1 - 5 were infected under non-life-threatening conditions for this experiment and are maintained as antibody donors for the following A. marginale strains as follows: Steer 1, Yautepec Mor.; steer 2, with Veracruz, Tizimín, and Yautepec strains; steers 3, 4, and 5 were infected with Tlapacoyan-1, Tlapacoyan-2 and Pt. de Ixtla strains, respectively. Steers 6 and 7 acquired infection under natural conditions and harbor Tlapacoyan-1 and Tlapacoyan-2 strains respectively; infected blood and serum samples from these two steers are kept under storage at -70 ºC.
Microorganisms
Nine Mexican A. marginale strains from different locations were used according to their differences in repeats in the Msp1a repeats15 as observed in Table 1.
Table 1 Mexican strains from which virB11 was sequenced
| Numeric ID | Denomination according to municipality and State | Msp1a variable repeats |
|---|---|---|
| Mex-28-037-01 | Soto la Marina, Tamaulipas | τ 57 13 18 |
| Mex-28-037-02 | Guayaboso, Tamaulipas | 28 29 74 29 M F |
| Mex-14-010-01 | Atitalaquia, Hidalgo | τ 57 13 18 |
| Mex-31-096-01 | Tizimín, Yucatán | T C B B C B π |
| Mex-31-089-01 | Ticul, Yucatán | F M M |
| Mex-30-184-03 | Tlapacoyan-2, Veracruz | 73 ( ( ( ( |
| Mex-30-130-01 | Playa Vicente, Veracruz | T C B B C B C |
| Mex-01-001-01 | Aguascalientes, Aguascalientes | 4 9 10 11 9 |
| Mex-17-017-01 | Puente de Ixtla, Morelos | 12 13 14 |
Anaplasma marginale strains were selected as for their geographic origin and differences in Msp1a repeats
Purification of Anaplasma marginale initial bodies
A. marginale initial bodies were purified from frozen bovine infected blood as described15 with modifications, briefly: once initial bodies were extruded form the erythrocytes, the material was separated through a 30 ml preformed linear 0-60% sucrose density gradient. 1.5 ml of the homogenized pellet was carefully layered on top of the preformed gradient and centrifuged at 7,000 xg for 30 min in a fixed angle rotor. After centrifugation, the intermediate phase containing the initial bodies (IB) was carefully recovered. IB were suspended in Puck’s saline in 30 mL tubes and centrifuged at 15,000 xg for 15 min at 4°C, twice in order to remove the sucrose. The supernatant was then removed and the pellet was suspended in 500 μl of Puck’s saline. The protein in the samples was quantitated by the Bradford method16, using bovine serum albumin (BSA) as standard. The IB were used immediately or stored at -70 °C, until use.
Cloning and sequencing of A. marginale virB11 gene
virb11 gene was amplified by PCR from genomic DNA (UltraClean® BloodSpin®DNA, MOBIO Laboratories Inc) obtained from cryopreserved (-70 ºC) blood, using Primers (F5’ GTGTTGTGTGCGCATATG 3’, R5’ CATTGCCTTGTGAACATTTAGTG 3’) under the following amplification program: 95 °C for 5 min, followed by 30 cycles at 95 °C for 45 sec, 56 °C for 1 min, 72 °C for 45 sec, with a final extension step at 72 °C for 10 min, using GoTaq® Green Master Mix 2X commercial mix (Promega, Madison, WI, USA). PCR products were separated by electrophoresis on 1% agarose gels and, as reference, the 1 kb Plus DNA Ladder (Invitrogen) weight marker was used; the image was captured with a KODAK Gel Logic 1500 System Picture system. PCR products were purified using a Wizard® SV Gel and PCR Clean-Up System commercial kit, and resuspended in 50 μl of water. One (1) μl of the product of each gene was cloned in a cloneJET PCR Cloning Kit (8878 Thermo-Scientific), and the mixture used for the transformation of competent Top10 E. coli cells. Plasmids from each gene were sequenced using T7 promoter forward and reverse primers, respectively. Sequencing was performed at the Biotechnology Institute, UNAM.
Bioinformatics analysis
The comparative identity among sequenced PCR products was determined with ClustalW17, using the putative virB11 gene of the A. marginale St. Maries strain as reference. The consensus sequences for each gene were translated into amino acids and used in the bioinformatics analysis. The prediction of the exposed extracellular domains was made with TMHMM program18. The ProtScale program was used for prediction of hydrophobic regions19 and the presence of linear type B epitopes was predicted with BCEPred, ABCPred and IEDB20,21 programs all available on-line.
Multiple antigenic peptide (MAP) design
Based on the analysis of linear Type B epitopes a multiple antigenic peptide was designed considering the highest scores for the following characteristics: antigenicity, hydrophobicity, flexibility, surface exposure and synthesis ease such as, sequence size and solubility of the present amino acids. MAPVirB11 was designed as a tetramer and its synthesis was commercially commissioned to American Peptide Company, Inc. (Fischer Scientific, USA).
MAPVirB11 is recognized by IgG of cattle infected with A. marginale by ELISA
MAPVirB11 tetramer antigenicity against A. marginale immune bovine sera was tested by indirect ELISA in 96-well microtiter plates (Corning 3595) coated with 100 μL/well of MAPVirB11 at a 10 μg/mL concentration, in carbonate buffer solution (CBS) pH 9.6; incubated overnight at 4 °C. Plates were washed three times with PBS pH 7.2 /0.05% Tween 20 (PBS/Tween), and blocked with 7% skim milk in PBS/Tween for 1 h at 37 °C; the contents was discarded, wells were washed, and probed with 100 μL/well of A. marginale immune bovine sera serially diluted from 1/25, up to 1/100. Antigen-antibody reaction was made visible by probing with anti-bovine IgG alkaline phosphatase conjugate (Phosphatase-Conjugated AffiniPure Goat Anti-Bovine, Jackson ImmunoResearch, Laboratories, Inc.) diluted at 1/5000. The plates were read on a BioRad iMark™ Microplate Absorbance Reader #1681135 at 405 nm.
Preparation of anti-MAP-VirB11 rabbit polyclonal monospecific serum
New Zealand rabbits were subcutaneously immunized weekly on five occasions at 7 d intervals; with 50 μg of MAPVirB11 tetramer diluted in 1 mL PBS and emulsified in 1 mL Montanide™ ISA 70 adjuvant. Before the first immunization (T0) serum was used as negative control for each rabbit. Serum samples were obtained on T0, and each immunization time T1, T2, T3, T4 and T5. Rabbit anti MAPVirB11 antibody specificity was determined by ELISA, as described above with modifications. 96-well microtiter plates (Corning 3595) were coated with 100 μL of MAPVirB11 with a concentration of 10 μg/mL in carbonate buffer. Rabbit serum samples were serially diluted from 1/25 to 1/100 in 0.05% PBS Tween. To detect the Ag-Ac reaction, was made visible with Anti-IgG-Rabbit Gtx Rbt IgG2 Alkaline Phosphate (Alka Phos) conjugate (Chemicon International; Cat. No. AP307P) was used and read as described.
Identification of native VirB11 in A. marginale by Western blot
Twenty (20) μg of total initial body (IB) protein of Tlapacoyan-2, A. marginale, were run on denaturing and reductive SDS-PAGE gels with a 3%/15% pre-separation/separation phases. Electrophoresis was performed at 150 V, at constant voltage for one hour. One replicate gel was washed for one hour in distilled water and stained in Coomassie GelCode ™ Blue Solution (Thermo Fisher Scientific).
The second replicate was transferred to a 0.45 μm pore (Immobilon®-P Millipore) PVDF membrane on a semi-dry system at 15 volts for 15 min. At the end of the transference, the membrane was blocked for 2 h with 5% skim milk in Tris buffered solution (TBS) pH 7.5 and washed three times with TBS-0.05% Tween (TBS Tween). The membrane was incubated at room temperature with a 1:100 dilution of T5 rabbit serum in TBS Tween for 2 h; after three washes the membrane was incubated with the Anti-IgG-Rabbit Gtx Rbt IgG2 Alka Phos diluted 1:5000 in PBS_0.05% Tween-20 for 2 h; (Chemicon International). The bands were detected by incubating the membrane with alkaline phosphatase substrate (BCIP / NBT SIGMA) for 5 to 20 min.
Rabbit monospecific polyclonal anti-MAPVirB11 antibodies react with bovine infected erythrocytes by immunofluorescence
Antigen slides prepared from a 15% rickettsemic steer (Puente de Ixtla strain), were air-dried and stored at -70 ºC. Ten (10) μL of rabbit T0 and T5 anti MAPVirB11, at a 1:50 dilution in PBS (pH 7.2), were placed on an antigen slide and incubated for 60 min as described22. Antigen slides were incubated at 37 °C for 30 min and washed three times with PBS, then, incubated with FITC-anti-rabbit antibody conjugate (Jackson Immuno Research Laboratories, Inc.). Slides were washed three times with PBS pH 7.4 and air-dried at room temperature and examined by epifluorescent microscopy.
Cell localization of VIRB11 in A. marginale by transmission electron microscopy
Density gradient-purified A. marginale initial bodies from Yautepec Strain (see supplemental material for extraction and purification of initial bodies) were placed on copper grids coated with Formvar 200 mesh (Senna Science) and incubated for 1 h at room temperature. After incubation, grids were washed three times with TBS and incubated with anti-MAP-VirB11 primary rabbit antibodies and a pre-immunization control serum in a 1:5 TBS dilution for 1 h. Grids were washed again three times for three minutes each, and incubated for 1 h with Sigma-Aldrich gold-labeled conjugated anti-rabbit secondary antibody (particle size of 10 nm). Grids were negatively stained with uranyl acetate and observed under a microscope at 4K, 8K and 20K (MET Microscopic, ZEISS, Libra 120 model with camera Gatan Multiscan Camera, 794 Model).
Results
Bioinformatics analysis
At amplification, each virB11 PCR product of the six Mexican A. marginale isolates showed an approximate 1,000 bp, molecular weight, consistent to that reported for the St. Marie strain. Nucleotide sequence alignments of nine Mexican isolates, showed 100 % identity among them (Table 2).
Table 2 virB11 gene was amplified and sequenced in nine Mexican A. marginale isolates
| St. Maries GTGTTGTGTGCGCATATGACAGCAGGATACGCAGCGTTAGAAACATACTTGGAGCCCCTG |
| Soto la Marina GTGTTGTGTGCGCATATGACAGCAGGATACGCAGCGTTAGAAACATACTTGGAGCCCCTG |
| Guayaboso GTGTTGTGTGCGCATATGACAGCAGGATACGCAGCGTTAGAAACATACTTGGAGCCCCTG |
| Atitalaquia GTGTTGTGTGCGCATATGACAGCAGGATACGCAGCGTTAGAAACATACTTGGAGCCCCTG |
| Tizimin GTGTTGTGTGCGCATATGACAGCAGGATACGCAGCGTTAGAAACATACTTGGAGCCCCTG |
| Ticul GTGTTGTGTGCGCATATGACAGCAGGATACGCAGCGTTAGAAACATACTTGGAGCCCCTG |
| Tlapacoyan-2 GTGTTGTGTGCGCATATGACAGCAGGATACGCAGCGTTAGAAACATACTTGGAGCCCCTG |
| Playa Vicente GTGTTGTGTGCGCATATGACAGCAGGATACGCAGCGTTAGAAACATACTTGGAGCCCCTG |
| Aguascalientes GTGTTGTGTGCGCATATGACAGCAGGATACGCAGCGTTAGAAACATACTTGGAGCCCCTG |
| Pte de Ixtla GTGTTGTGTGCGCATATGACAGCAGGATACGCAGCGTTAGAAACATACTTGGAGCCCCTG |
| ************************************************************ |
| St. Maries CAGAGCATATTCGCGGAGGATGGGGTCAATGAGATCTCGATCAACAGAGAATGCGAGGTA |
| Soto la Marina CAGAGCATATTCGCGGAGGATGGGGTCAATGAGATCTCGATCAACAGAGAATGCGAGGTA |
| Guayaboso CAGAGCATATTCGCGGAGGATGGGGTCAATGAGATCTCGATCAACAGAGAATGCGAGGTA |
| Atitalaquia CAGAGCATATTCGCGGAGGATGGGGTCAATGAGATCTCGATCAACAGAGAATGCGAGGTA |
| Tizimin CAGAGCATATTCGCGGAGGATGGGGTCAATGAGATCTCGATCAACAGAGAATGCGAGGTA |
| Ticul CAGAGCATATTCGCGGAGGATGGGGTCAATGAGATCTCGATCAACAGAGAATGCGAGGTA |
| Tlapacoyan-2 CAGAGCATATTCGCGGAGGATGGGGTCAATGAGATCTCGATCAACAGAGAATGCGAGGTA |
| Playa Vicente CAGAGCATATTCGCGGAGGATGGGGTCAATGAGATCTCGATCAACAGAGAATGCGAGGTA |
| Aguascalientes CAGAGCATATTCGCGGAGGATGGGGTCAATGAGATCTCGATCAACAGAGAATGCGAGGTA |
| Pte de Ixtla CAGAGCATATTCGCGGAGGATGGGGTCAATGAGATCTCGATCAACAGAGAATGCGAGGTA |
| ************************************************************ |
| St. Maries TGGGTAGAGAACAGGGGCGATATACGGTGTGAGAGGATTGAAAGTCTAACTCTGTCGCAT |
| Soto la Marina TGGGTAGAGAACAGGGGCGATATACGGTGTGAGAGGATTGAAAGTCTAACTCTGTCGCAT |
| Guayaboso TGGGTAGAGAACAGGGGCGATATACGGTGTGAGAGGATTGAAAGTCTAACTCTGTCGCAT |
| Atitalaquia TGGGTAGAGAACAGGGGCGATATACGGTGTGAGAGGATTGAAAGTCTAACTCTGTCGCAT |
| Tizimin TGGGTAGAGAACAGGGGCGATATACGGTGTGAGAGGATTGAAAGTCTAACTCTGTCGCAT |
| Ticul TGGGTAGAGAACAGGGGCGATATACGGTGTGAGAGGATTGAAAGTCTAACTCTGTCGCAT |
| Tlapacoyan-2 TGGGTAGAGAACAGGGGCGATATACGGTGTGAGAGGATTGAAAGTCTAACTCTGTCGCAT |
| Playa Vicente TGGGTAGAGAACAGGGGCGATATACGGTGTGAGAGGATTGAAAGTCTAACTCTGTCGCAT |
| Aguascalientes TGGGTAGAGAACAGGGGCGATATACGGTGTGAGAGGATTGAAAGTCTAACTCTGTCGCAT |
| Pte de Ixtla TGGGTAGAGAACAGGGGCGATATACGGTGTGAGAGGATTGAAAGTCTAACTCTGTCGCAT |
| ************************************************************ |
| St. Maries CTAAAAGCACTCGGCAGGTTGGTAGCACAAGCAACCGAGCAGAAATTGAGTGAGGAAACG |
| Soto la Marina CTAAAAGCACTCGGCAGGTTGGTAGCACAAGCAACCGAGCAGAAATTGAGTGAGGAAACG |
| Guayaboso CTAAAAGCACTCGGCAGGTTGGTAGCACAAGCAACCGAGCAGAAATTGAGTGAGGAAACG |
| Atitalaquia CTAAAAGCACTCGGCAGGTTGGTAGCACAAGCAACCGAGCAGAAATTGAGTGAGGAAACG |
| Tizimin CTAAAAGCACTCGGCAGGTTGGTAGCACAAGCAACCGAGCAGAAATTGAGTGAGGAAACG |
| Ticul CTAAAAGCACTCGGCAGGTTGGTAGCACAAGCAACCGAGCAGAAATTGAGTGAGGAAACG |
| Tlapacoyan-2 CTAAAAGCACTCGGCAGGTTGGTAGCACAAGCAACCGAGCAGAAATTGAGTGAGGAAACG |
| Playa Vicente CTAAAAGCACTCGGCAGGTTGGTAGCACAAGCAACCGAGCAGAAATTGAGTGAGGAAACG |
| Aguascalientes CTAAAAGCACTCGGCAGGTTGGTAGCACAAGCAACCGAGCAGAAATTGAGTGAGGAAACG |
| Pte de Ixtla CTAAAAGCACTCGGCAGGTTGGTAGCACAAGCAACCGAGCAGAAATTGAGTGAGGAAACG |
| ************************************************************ |
| St. Maries CCTTTGCTGTCGGCTTCTTTGCCCAACGGCTTCCGCGTGCAGGTGGTGTTTCCCCCCGCT |
| Soto la Marina CCTTTGCTGTCGGCTTCTTTGCCCAACGGCTTCCGCGTGCAGGTGGTGTTTCCCCCCGCT |
| Guayaboso CCTTTGCTGTCGGCTTCTTTGCCCAACGGCTTCCGCGTGCAGGTGGTGTTTCCCCCCGCT |
| Atitalaquia CCTTTGCTGTCGGCTTCTTTGCCCAACGGCTTCCGCGTGCAGGTGGTGTTTCCCCCCGCT |
| Tizimin CCTTTGCTGTCGGCTTCTTTGCCCAACGGCTTCCGCGTGCAGGTGGTGTTTCCCCCCGCT |
| Ticul CCTTTGCTGTCGGCTTCTTTGCCCAACGGCTTCCGCGTGCAGGTGGTGTTTCCCCCCGCT |
| Tlapacoyan-2 CCTTTGCTGTCGGCTTCTTTGCCCAACGGCTTCCGCGTGCAGGTGGTGTTTCCCCCCGCT |
| Playa Vicente CCTTTGCTGTCGGCTTCTTTGCCCAACGGCTTCCGCGTGCAGGTGGTGTTTCCCCCCGCT |
| Aguascalientes CCTTTGCTGTCGGCTTCTTTGCCCAACGGCTTCCGCGTGCAGGTGGTGTTTCCCCCCGCT |
| Pte de Ixtla CCTTTGCTGTCGGCTTCTTTGCCCAACGGCTTCCGCGTGCAGGTGGTGTTTCCCCCCGCT |
| ************************************************************ |
| St. Maries TGCGAGGGTGATAAAGTCGTATTTTCCATAAGAAAGCCATCGACGGTGCAGTTGTCGCTC |
| Soto la Marina TGCGAGGGTGATAAAGTCGTATTTTCCATAAGAAAGCCATCGACGGTGCAGTTGTCGCTC |
| Guayaboso TGCGAGGGTGATAAAGTCGTATTTTCCATAAGAAAGCCATCGACGGTGCAGTTGTCGCTC |
| Atitalaquia TGCGAGGGTGATAAAGTCGTATTTTCCATAAGAAAGCCATCGACGGTGCAGTTGTCGCTC |
| Tizimin TGCGAGGGTGATAAAGTCGTATTTTCCATAAGAAAGCCATCGACGGTGCAGTTGTCGCTC |
| Ticul TGCGAGGGTGATAAAGTCGTATTTTCCATAAGAAAGCCATCGACGGTGCAGTTGTCGCTC |
| Tlapacoyan-2 TGCGAGGGTGATAAAGTCGTATTTTCCATAAGAAAGCCATCGACGGTGCAGTTGTCGCTC |
| Playa Vicente TGCGAGGGTGATAAAGTCGTATTTTCCATAAGAAAGCCATCGACGGTGCAGTTGTCGCTC |
| Aguascalientes TGCGAGGGTGATAAAGTCGTATTTTCCATAAGAAAGCCATCGACGGTGCAGTTGTCGCTC |
| Pte de Ixtla TGCGAGGGTGATAAAGTCGTATTTTCCATAAGAAAGCCATCGACGGTGCAGTTGTCGCTC |
| ************************************************************ |
| St. Maries GATGATTATGAAAAAATGGGTGCGTTCTCCCATGTGGCCCAGCAGGGCCGCAAGGCTATG |
| Soto la Marina GATGATTATGAAAAAATGGGTGCGTTCTCCCATGTGGCCCAGCAGGGCCGCAAGGCTATG |
| Guayaboso GATGATTATGAAAAAATGGGTGCGTTCTCCCATGTGGCCCAGCAGGGCCGCAAGGCTATG |
| Atitalaquia GATGATTATGAAAAAATGGGTGCGTTCTCCCATGTGGCCCAGCAGGGCCGCAAGGCTATG |
| Tizimin GATGATTATGAAAAAATGGGTGCGTTCTCCCATGTGGCCCAGCAGGGCCGCAAGGCTATG |
| Ticul GATGATTATGAAAAAATGGGTGCGTTCTCCCATGTGGCCCAGCAGGGCCGCAAGGCTATG |
| Tlapacoyan GATGATTATGAAAAAATGGGTGCGTTCTCCCATGTGGCCCAGCAGGGCCGCAAGGCTATG |
| Playa Vicente GATGATTATGAAAAAATGGGTGCGTTCTCCCATGTGGCCCAGCAGGGCCGCAAGGCTATG |
| Aguascalientes GATGATTATGAAAAAATGGGTGCGTTCTCCCATGTGGCCCAGCAGGGCCGCAAGGCTATG |
| Puente GATGATTATGAAAAAATGGGTGCGTTCTCCCATGTGGCCCAGCAGGGCCGCAAGGCTATG |
| ************************************************************ |
| St. Maries GACGTAAACAAGCAGCTTAGTGAGCTCCTGGATAGCGGGGATATCAAATCTTTCATAGAG |
| Soto la Marina GACGTAAACAAGCAGCTTAGTGAGCTCCTGGATAGCGGGGATATCAAATCTTTCATAGAG |
| Guayaboso GACGTAAACAAGCAGCTTAGTGAGCTCCTGGATAGCGGGGATATCAAATCTTTCATAGAG |
| Atitalaquia GACGTAAACAAGCAGCTTAGTGAGCTCCTGGATAGCGGGGATATCAAATCTTTCATAGAG |
| Tizimin GACGTAAACAAGCAGCTTAGTGAGCTCCTGGATAGCGGGGATATCAAATCTTTCATAGAG |
| Ticul GACGTAAACAAGCAGCTTAGTGAGCTCCTGGATAGCGGGGATATCAAATCTTTCATAGAG |
| Tlapacoyan-2 GACGTAAACAAGCAGCTTAGTGAGCTCCTGGATAGCGGGGATATCAAATCTTTCATAGAG |
| Playa Vicente GACGTAAACAAGCAGCTTAGTGAGCTCCTGGATAGCGGGGATATCAAATCTTTCATAGAG |
| Aguascalientes GACGTAAACAAGCAGCTTAGTGAGCTCCTGGATAGCGGGGATATCAAATCTTTCATAGAG |
| Puente GACGTAAACAAGCAGCTTAGTGAGCTCCTGGATAGCGGGGATATCAAATCTTTCATAGAG |
| ************************************************************ |
| St. Maries CTTGCGGTGCTGAGCAAAAAAAACATAATCGTCAGTGGAGGTACGTCCACTGGGAAAACT |
| Soto la Marina CTTGCGGTGCTGAGCAAAAAAAACATAATCGTCAGTGGAGGTACGTCCACTGGGAAAACT |
| Guayaboso CTTGCGGTGCTGAGCAAAAAAAACATAATCGTCAGTGGAGGTACGTCCACTGGGAAAACT |
| Atitalaquia CTTGCGGTGCTGAGCAAAAAAAACATAATCGTCAGTGGAGGTACGTCCACTGGGAAAACT |
| Tizimin CTTGCGGTGCTGAGCAAAAAAAACATAATCGTCAGTGGAGGTACGTCCACTGGGAAAACT |
| Ticul CTTGCGGTGCTGAGCAAAAAAAACATAATCGTCAGTGGAGGTACGTCCACTGGGAAAACT |
| Tlapacoyan-2 CTTGCGGTGCTGAGCAAAAAAAACATAATCGTCAGTGGAGGTACGTCCACTGGGAAAACT |
| Playa Vicente CTTGCGGTGCTGAGCAAAAAAAACATAATCGTCAGTGGAGGTACGTCCACTGGGAAAACT |
| Aguascalientes CTTGCGGTGCTGAGCAAAAAAAACATAATCGTCAGTGGAGGTACGTCCACTGGGAAAACT |
| Puente CTTGCGGTGCTGAGCAAAAAAAACATAATCGTCAGTGGAGGTACGTCCACTGGGAAAACT |
| ************************************************************ |
| St. Maries ACGTTCACAAACGCGGCGCTGCGGGTCATCCCAAAGGATGAAAGGATAATAACGGTGGAA |
| Soto la Marina ACGTTCACAAACGCGGCGCTGCGGGTCATCCCAAAGGATGAAAGGATAATAACGGTGGAA |
| Guayaboso ACGTTCACAAACGCGGCGCTGCGGGTCATCCCAAAGGATGAAAGGATAATAACGGTGGAA |
| Atitalaquia ACGTTCACAAACGCGGCGCTGCGGGTCATCCCAAAGGATGAAAGGATAATAACGGTGGAA |
| Tizimin ACGTTCACAAACGCGGCGCTGCGGGTCATCCCAAAGGATGAAAGGATAATAACGGTGGAA |
| Ticul ACGTTCACAAACGCGGCGCTGCGGGTCATCCCAAAGGATGAAAGGATAATAACGGTGGAA |
| Tlapacoyan-2 ACGTTCACAAACGCGGCGCTGCGGGTCATCCCAAAGGATGAAAGGATAATAACGGTGGAA |
| Playa Vicente ACGTTCACAAACGCGGCGCTGCGGGTCATCCCAAAGGATGAAAGGATAATAACGGTGGAA |
| Aguascalientes ACGTTCACAAACGCGGCGCTGCGGGTCATCCCAAAGGATGAAAGGATAATAACGGTGGAA |
| Pte de Ixtla ACGTTCACAAACGCGGCGCTGCGGGTCATCCCAAAGGATGAAAGGATAATAACGGTGGAA |
| ************************************************************ |
| St. Maries GACTCTAGAGAAATAGCGCTAGACCACCCCAACAGGGTACATCTATTGGCATCCAAGGGC |
| Soto la Marina GACTCTAGAGAAATAGCGCTAGACCACCCCAACAGGGTACATCTATTGGCATCCAAGGGC |
| Guayaboso GACTCTAGAGAAATAGCGCTAGACCACCCCAACAGGGTACATCTATTGGCATCCAAGGGC |
| Atitalaquia GACTCTAGAGAAATAGCGCTAGACCACCCCAACAGGGTACATCTATTGGCATCCAAGGGC |
| Tizimin GACTCTAGAGAAATAGCGCTAGACCACCCCAACAGGGTACATCTATTGGCATCCAAGGGC |
| Ticul GACTCTAGAGAAATAGCGCTAGACCACCCCAACAGGGTACATCTATTGGCATCCAAGGGC |
| Tlapacoyan-2 GACTCTAGAGAAATAGCGCTAGACCACCCCAACAGGGTACATCTATTGGCATCCAAGGGC |
| Playa Vicente GACTCTAGAGAAATAGCGCTAGACCACCCCAACAGGGTACATCTATTGGCATCCAAGGGC |
| Aguascalientes GACTCTAGAGAAATAGCGCTAGACCACCCCAACAGGGTACATCTATTGGCATCCAAGGGC |
| Pte de Ixtla GACTCTAGAGAAATAGCGCTAGACCACCCCAACAGGGTACATCTATTGGCATCCAAGGGC |
| ************************************************************ |
| St. Maries GGCCAGGGGCGCGCTAAGGTCTCTACTCAAGATCTTATAGAGGCCTGCTTGCGTCTTCGT |
| Soto la Marina GGCCAGGGGCGCGCTAAGGTCTCTACTCAAGATCTTATAGAGGCCTGCTTGCGTCTTCGT |
| Guayaboso GGCCAGGGGCGCGCTAAGGTCTCTACTCAAGATCTTATAGAGGCCTGCTTGCGTCTTCGT |
| Atitalaquia GGCCAGGGGCGCGCTAAGGTCTCTACTCAAGATCTTATAGAGGCCTGCTTGCGTCTTCGT |
| Tizimin GGCCAGGGGCGCGCTAAGGTCTCTACTCAAGATCTTATAGAGGCCTGCTTGCGTCTTCGT |
| Ticul GGCCAGGGGCGCGCTAAGGTCTCTACTCAAGATCTTATAGAGGCCTGCTTGCGTCTTCGT |
| Tlapacoyan-2 GGCCAGGGGCGCGCTAAGGTCTCTACTCAAGATCTTATAGAGGCCTGCTTGCGTCTTCGT |
| Playa Vicente GGCCAGGGGCGCGCTAAGGTCTCTACTCAAGATCTTATAGAGGCCTGCTTGCGTCTTCGT |
| Aguascalientes GGCCAGGGGCGCGCTAAGGTCTCTACTCAAGATCTTATAGAGGCCTGCTTGCGTCTTCGT |
| Pte de Ixtla GGCCAGGGGCGCGCTAAGGTCTCTACTCAAGATCTTATAGAGGCCTGCTTGCGTCTTCGT |
| ************************************************************ |
| St. Maries CCAGATAGAATAATTGTTGGGGAGCTCAGGGGGGCTGAGGCGTTTAGCTTTCTCAGAGCC |
| Soto la Marina CCAGATAGAATAATTGTTGGGGAGCTCAGGGGGGCTGAGGCGTTTAGCTTTCTCAGAGCC |
| Guayaboso CCAGATAGAATAATTGTTGGGGAGCTCAGGGGGGCTGAGGCGTTTAGCTTTCTCAGAGCC |
| Atitalaquia CCAGATAGAATAATTGTTGGGGAGCTCAGGGGGGCTGAGGCGTTTAGCTTTCTCAGAGCC |
| Tizimin CCAGATAGAATAATTGTTGGGGAGCTCAGGGGGGCTGAGGCGTTTAGCTTTCTCAGAGCC |
| Ticul CCAGATAGAATAATTGTTGGGGAGCTCAGGGGGGCTGAGGCGTTTAGCTTTCTCAGAGCC |
| Tlapacoyan-2 CCAGATAGAATAATTGTTGGGGAGCTCAGGGGGGCTGAGGCGTTTAGCTTTCTCAGAGCC |
| Playa Vicente CCAGATAGAATAATTGTTGGGGAGCTCAGGGGGGCTGAGGCGTTTAGCTTTCTCAGAGCC |
| Aguascalientes CCAGATAGAATAATTGTTGGGGAGCTCAGGGGGGCTGAGGCGTTTAGCTTTCTCAGAGCC |
| Pte de Ixtla CCAGATAGAATAATTGTTGGGGAGCTCAGGGGGGCTGAGGCGTTTAGCTTTCTCAGAGCC |
| ************************************************************ |
| St. Maries ATAAATACTGGGCACCCGGGTTCCATCTCAACCTTGCATGCAGACACGCCTAGAATGGCT |
| Soto la Marina ATAAATACTGGGCACCCGGGTTCCATCTCAACCTTGCATGCAGACACGCCTAGAATGGCT |
| Guayaboso ATAAATACTGGGCACCCGGGTTCCATCTCAACCTTGCATGCAGACACGCCTAGAATGGCT |
| Atitalaquia ATAAATACTGGGCACCCGGGTTCCATCTCAACCTTGCATGCAGACACGCCTAGAATGGCT |
| Tizimin ATAAATACTGGGCACCCGGGTTCCATCTCAACCTTGCATGCAGACACGCCTAGAATGGCT |
| Ticul ATAAATACTGGGCACCCGGGTTCCATCTCAACCTTGCATGCAGACACGCCTAGAATGGCT |
| Tlapacoyan-2 ATAAATACTGGGCACCCGGGTTCCATCTCAACCTTGCATGCAGACACGCCTAGAATGGCT |
| Playa Vicente ATAAATACTGGGCACCCGGGTTCCATCTCAACCTTGCATGCAGACACGCCTAGAATGGCT |
| Aguascalientes ATAAATACTGGGCACCCGGGTTCCATCTCAACCTTGCATGCAGACACGCCTAGAATGGCT |
| Pte de Ixtla ATAAATACTGGGCACCCGGGTTCCATCTCAACCTTGCATGCAGACACGCCTAGAATGGCT |
| ************************************************************ |
| St. Maries GTAGAGCAGCTGAAACTTATGGTAATGCAAGCGAGTCTAGGGTTGCCGCCAGACCAAATA |
| Soto la Marina GTAGAGCAGCTGAAACTTATGGTAATGCAAGCGAGTCTAGGGTTGCCGCCAGACCAAATA |
| Guayaboso GTAGAGCAGCTGAAACTTATGGTAATGCAAGCGAGTCTAGGGTTGCCGCCAGACCAAATA |
| Atitalaquia GTAGAGCAGCTGAAACTTATGGTAATGCAAGCGAGTCTAGGGTTGCCGCCAGACCAAATA |
| Tizimin GTAGAGCAGCTGAAACTTATGGTAATGCAAGCGAGTCTAGGGTTGCCGCCAGACCAAATA |
| Ticul GTAGAGCAGCTGAAACTTATGGTAATGCAAGCGAGTCTAGGGTTGCCGCCAGACCAAATA |
| Tlapacoyan-2 GTAGAGCAGCTGAAACTTATGGTAATGCAAGCGAGTCTAGGGTTGCCGCCAGACCAAATA |
| Playa Vicente GTAGAGCAGCTGAAACTTATGGTAATGCAAGCGAGTCTAGGGTTGCCGCCAGACCAAATA |
| Aguascalientes GTAGAGCAGCTGAAACTTATGGTAATGCAAGCGAGTCTAGGGTTGCCGCCAGACCAAATA |
| Pte de Ixtla GTAGAGCAGCTGAAACTTATGGTAATGCAAGCGAGTCTAGGGTTGCCGCCAGACCAAATA |
| ************************************************************ |
| St. Maries GTAAGTTACATTACCAACATAGTAGACGTAATTATACAGCTGAAAAGGGAGTCTGGAGGG |
| Soto la Marina GTAAGTTACATTACCAACATAGTAGACGTAATTATACAGCTGAAAAGGGAGTCTGGAGGG |
| Guayaboso GTAAGTTACATTACCAACATAGTAGACGTAATTATACAGCTGAAAAGGGAGTCTGGAGGG |
| Atitalaquia GTAAGTTACATTACCAACATAGTAGACGTAATTATACAGCTGAAAAGGGAGTCTGGAGGG |
| Tizimin GTAAGTTACATTACCAACATAGTAGACGTAATTATACAGCTGAAAAGGGAGTCTGGAGGG |
| Ticul GTAAGTTACATTACCAACATAGTAGACGTAATTATACAGCTGAAAAGGGAGTCTGGAGGG |
| Tlapacoyan-2 GTAAGTTACATTACCAACATAGTAGACGTAATTATACAGCTGAAAAGGGAGTCTGGAGGG |
| Playa Vicente GTAAGTTACATTACCAACATAGTAGACGTAATTATACAGCTGAAAAGGGAGTCTGGAGGG |
| Aguascalientes GTAAGTTACATTACCAACATAGTAGACGTAATTATACAGCTGAAAAGGGAGTCTGGAGGG |
| Pte de Ixtla GTAAGTTACATTACCAACATAGTAGACGTAATTATACAGCTGAAAAGGGAGTCTGGAGGG |
| ************************************************************ |
| St. Maries GTGCGGCACGTCGCGGAGATTATGTTCACTAAATGTTCACAAGGCAATGATTAA |
| Soto la Marina GTGCGGCACGTCGCGGAGATTATGTTCACTAAATGTTCACAAGGCAATGATTAA |
| Guayaboso GTGCGGCACGTCGCGGAGATTATGTTCACTAAATGTTCACAAGGCAATGATTAA |
| Atitalaquia GTGCGGCACGTCGCGGAGATTATGTTCACTAAATGTTCACAAGGCAATGATTAA |
| Tizimin GTGCGGCACGTCGCGGAGATTATGTTCACTAAATGTTCACAAGGCAATGATTAA |
| Ticul GTGCGGCACGTCGCGGAGATTATGTTCACTAAATGTTCACAAGGCAATGATTAA |
| Tlapacoyan-2 GTGCGGCACGTCGCGGAGATTATGTTCACTAAATGTTCACAAGGCAATGATTAA |
| Playa Vicente GTGCGGCACGTCGCGGAGATTATGTTCACTAAATGTTCACAAGGCAATGATTAA |
| Aguascalientes GTGCGGCACGTCGCGGAGATTATGTTCACTAAATGTTCACAAGGCAATGATTAA |
| Pte de Ixtla GTGCGGCACGTCGCGGAGATTATGTTCACTAAATGTTCACAAGGCAATGATTAA |
| ****************************************************** |
The computational inference analyzes of the consensus sequences showed two hydrophobic regions, absence of signal peptide and the probable external localization on the membrane, according to the amino acid analysis.
VirB11 protein type B epitope analysis and selection of relevant epitope
B type epitope analysis showed that VFSIRKPSTVQLSD amino acid sequence filled the criteria for immunogenicity, ease of synthesis and used for tetramer design. This sequence is located at the 107 to 122 positions, within the hydrophobic extramembranal region of the protein. This peptide sequence showed the highest scores as a linear type B epitope in the BCpred, ACBpred and IEDB prediction programs (Table 3).
Sera of cattle naturally or experimentally infected with A. marginale reacts with MAPVirB11
Serum samples of seven cattle naturally or experimentally infected with A. marginale gave a response above the negative control absorbance value, some reaching ≥2 OD values at ELISA (Figure 1A). The serum from a susceptible animal, serologically and PCR negative to A. marginale was included as negative control, which showed a 0.5 OD value. In order to confirm these results, the sera from bovines 1 and 3, were tested by western blot (Figure 1B). Both sera reacted with a 6 kDa band, which coincides with the molecular weight of MAP-VirB11.
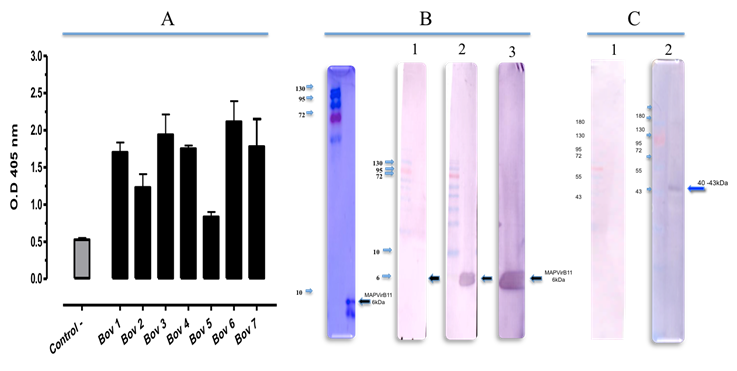
Figure 1 A) MAP-VirB11 reacts with bovine immune serum in ELISA: Sera from seven A. marginale-infected bovines were tested. The description of the animals is as above in Materials and Methods; B) Infected bovines serum reacts with Map-VirB11 in western blot. MAP-VirB11 was separated by SDS-PAGE and blotted onto PVDF membrane and probed with 1). A. marginale negative bovine serum; 2) Bovine 1 serum or 3) Bovine 3 serum. C) Rabbit T5 anti-MAP-VirB11 reacts with native de VirB11; purified A. marginale initial bodies were separated on a 15% polyacrylamide gel and transferred to a PVDF membrane and probed with: 1, non-immune rabbit serum; 2, immune Rabbit T5 serum.
Rabbit specific anti MAP-VirB11 antibody react with A. marginale native VirB11
Rabbit anti-MAPVirB11 antibodies reacted with MAPVirB11 in an ELISA (data not shown). All three sera reacted with similar OD values ≥ 0.3 to 0.5, which were higher than the one obtained for the control serum; the negative control had an OD value less than 0.1. In order to verify the rabbit immune serum specificity, rabbit antibodies were tested by western blot against the tetramer and proteins of the purified initial bodies. Figure 1C shows (lane 2) recognition of a 40 kDa band by T5 immune sera from the same rabbit whereas T0 non immune serum shows no signal.
Rabbit immune anti-MAPVirB11 serum reacts with intraerythrocytic A. marginale by immunofluorescence
Preimmune (T0) and immune sera (T5) were used to test if the immune anti MAPVirB11 antibodies reacted with intra-erythrocytic A. marginale in a direct immunofluorescence assay. Blood-smears of both non-infected (Figure 2A and 2B) and erythrocytes infected with Tlapacoyan-2 strain (2C and 2D). Smears were probed with both T0 (2A and 2C) and T5 (2B and 2D) immune sera. Figure 2D shows typical intraerythrocytic A. marginale bodies by direct fluorescence, in contrast to T0 serum, which shows no reaction (Figure 2C).
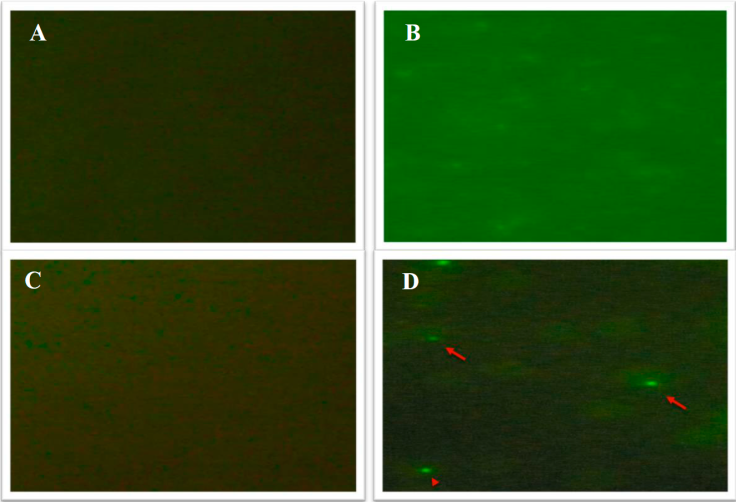
Figure 2 Recognition of intraerythrocytic A. marginale by rabbit immune serum anti-MAPVirB11 by direct immunofluorescence. Pre-immune rabbit serum was tested against A) non-infected erythrocytes or; B) infected erythrocytes; T5 rabbit anti-MAPVirB11 immune serum was tested against C) non-infected erythrocytes, and D) infected erythrocytes (100X magnification).
Cellular localization of VirB11 protein in A. marginale by transmission electron microscopy (TEM)
In order to determine the location of VirB11 native protein in A. marginale, purified initial bodies were probed with rabbit pre-immune T0 and immune T5 anti-MAPVirB11 sera and a second anti-rabbit IgG antibody coupled with colloidal gold. Figure 3B shows that T5 rabbit anti-MAPVirB11 antibodies recognized the VirB11 native protein on the surface of A. marginale, located within an associated region on the outer side of the rickettsial membrane (indicated by arrows). This recognition can be observed in more detail in Figures 3B1 and 3B2. Figure 3A shows the initial body of A. marginale probed with pre-immune serum as negative control. Additionally, it was evaluated the T5 rabbit anti-MAP antibodies against E. coli, which is not known to express the VirB11 protein (Figure 3C). No recognition was observed in this case (Figures 3C1), which is evidence of the anti-MAPVirB11 specificity (Figure 3B).
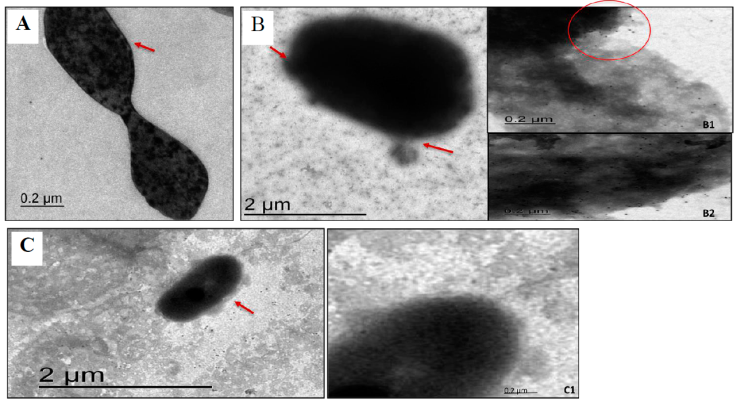
Figure 3 Rabbit anti-MAPVirB11 immune serum reacts with the outer membrane of A. marginale initial bodies by Transmission electron microscopy. A) Pre-immune rabbit serum incubated with A. marginale initial bodies, (A1 4K and A2 8K; B) MAPVirB11 specific rabbit serum incubated with A. marginale initial bodies (B1 and B2 are magnification views from image B); C) MAPVirB11 specific rabbit serum incubated with E. coli as control (C1 is a magnification view from image C).
Discussion
The publication of the complete genome sequence of the St. Maries strain of A. marginale23, opened a new era in the search for potential vaccine candidates against A. marginale. The possibility of these antigens to be used as immunogens under homologous challenge/vaccination conditions has been promising, however, immunity has not yet met the expectations under conditions of heterologous challenge as a consequence of the variability of the selected antigens and lack on information in a broader array of A. marginale strains. VirB11 a component of T4SS, has been attributed an ATPase function in other organisms.
Multiple alignment analysis of nine VirB11 nucleotide and amino acid sequences from Mexican strains showed 100 % identity (Figure 1) among themselves and with those reported in GenBank for the VirB11 of several strains including the St. Maries strain. The bioinformatic analysis also indicated that the protein was exposed on the outer surface of the membrane. Furthermore, 6 hydrophilic linear type B epitopes identified are conserved in all strains and exposed in the extracellular domain (Table 1); from these, we selected VFSIRKPSTVQLSLD amino acid sequence, which showed the highest weight of the aforementioned characteristics for the synthesis of MAP-VirB11.
In order to verify the possibility that infected animals produce antibodies against this protein, serum samples from naturally and experimentally infected cattle with A. marginale reacted, in an antigen-antibody recognition assay, against MAPVirB11 epitope (Figure 2A). This result is consistent with previous reports24 where bovines immunized with experimentally cross-linked membrane fragments from initial bodies of St. Maries strain induced an antibody response against VirB11 and other proteins of the same complex.
In order to determine the antigenicity of MAPVirB11, it was important to prove that the specific rabbit antibodies against MAP-VirB11 could recognize the native protein. This was corroborated; monospecific polyclonal rabbit antibodies induced by immunization with MAPVirB11 reacted in a specific manner when tested by ELISA. Similarly, sera from infected cattle reacted with a 6 kDa band, corresponding to the weight of MAPVirB11 by western blot (Figure 2B).
An important condition for a protein or peptide, to be considered as a potentially useful immunogen, is that it has to be exposed in the outer membrane of the rickettsia. Antibodies belonging to one rabbit immunized with MAPVirB11 and evaluated by indirect immunofluorescence, allowed us to identify A. marginale in the marginal zone of the infected erythrocytes; confirming that the antibodies induced by peptide inoculation, were capable of reacting with native VirB11 present in A. marginale (Figure 2).
Through the use of transmission electron microscopy and the same rabbit serum previously evaluated in WB and IFI assays, we identified the location of the VirB11 protein on the rickettsia; noting the labeling presence on a membrane section that was covering the periphery of the initial bodies of A. marginale, which in turn were extracted from infected erythrocytes (Figure 3). This evidence indicates that the protein is exposed on the Anaplasma outer surface, and is likely to be associated with the invasion process or survival within the erythrocyte25. This finding is in agreement with similar results regarding the localization of the VirB9 in A. phagocytophilum and its possible participation in the invasion mechanism of this organism in human neutrophils26.
Our strategy of selecting MAP’s within the immunogen design allowed us to direct the immune response and induced the molecular recognition towards one of the most interesting antigens of A. marginale. This highly versatile tool provided a specific response and a higher sensitivity in the detection system, placing it in a superior category compared to previous strategies. These findings show that MAP’s may be considered in the design of future diagnostic and immunization tools, for controlling of bovine anaplasmosis.
Conclusions and implications
It was demonstrated the presence of linear epitopes of the VirB11 protein conserved among nine Mexican strains of A. marginale, using bioinformatic analyses. Also, a tetramer peptide (MAP’s) designed based on the B-type epitope amino acid VFSIRKPSTVQLSLD VirB11 sequence; sera from infected cattle recognized this tetramer. Furthermore, specific rabbit anti-tetramer antibodies recognized VirB11 native protein in A. marginale-infected erythrocytes both by immunofluorescence and transmission electron microscopy. All this evidence, suggests that VirB11 may have potential as an important target of the bovine immune system. This study also demonstrated the usefulness of a tetramer (MAP) for diagnosis and generation of the immune response against native proteins of interest. Clearly, further studies are required to evaluate the ability of these molecular constructs in the control the A. marginale infection in cattle.
Ethics Statement
Two month-old New Zealand rabbits were kept in the Laboratory Animal Facility of the National Institute of Public Health, Cuernavaca, Mor., under the guidelines of the Institutional Committee for the Care and Use of Experimental Animals (SICUAE) of the National Autonomous University of Mexico, under Protocol No. DC-2013/1-3.
Competing interests/Funding Sources
The Mexican National Council for Science and Technology (CONACyT), and the National Institute for Research on Agriculture, Forestry and Livestock (INIFAP) funded this work under agreements No. 168167 and SIGI 1320382022 respectively.
The funding sources provided financial support for the conduction of the research and preparation of the manuscript. The content of the manuscript however is the authors’ sole responsibility, solely to acknowledge this fact.
This work is part of the requirements for graduation of the first author América Ivette Barrera Molina











 nueva página del texto (beta)
nueva página del texto (beta)


