Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO
Share
Revista mexicana de ciencias pecuarias
On-line version ISSN 2448-6698Print version ISSN 2007-1124
Rev. mex. de cienc. pecuarias vol.6 n.4 Mérida Oct./Dec. 2015
Articles
Loci associated with genetic disorders and meat quality in Charolais cattle in Mexico
a Laboratorio de Biotecnología Animal, Centro de Biotecnología Genómica. Instituto Politécnico Nacional. Boulevard del Maestro esq. Elías Piña S/N, Col. Narciso Mendoza, 88710 Cd. Reynosa, Tamaulipas, México. Tel: (899) 924 36 27.
b Charolais Herd Book de México A.C. Comité Técnico. México.
Allelic and genotypic frequencies of eight markers previously associated to genetic disorders and meat quality and located in the calpain (CAPN1-4751 and CAPN1-316), calpastatin (CAST-T1), thyroglobulin (TG5), myostatin (MSTN, Q204X), argininosuccinate synthase (ASS), monophosphate synthase (UMPS) and myophosphorylase (PYGM) genes, were determined from 493 registered Charolais animals sampled from two herds located at Sonora (n= 157) and three at Nuevo León (n= 336), Mexico. No carriers of mutated alleles of ASS and UMPS genes were found, but carriers of the MSTN Q204X allele were identified at frequencies of ≤ 1 % in Sonora populations and 8.6 to 14.4 % in Nuevo Leon. In addition, carriers of PYGM marker were identified at frequencies of 6.49 and 1% in a herd from Sonora and other from Nuevo León, respectively. Analysis of gene differentiation among herds and with four loci showed that there are highly significant differences within Northwest populations (P<0.0001) and between them and the Northeast (P<0.001), differentiation is mainly explained by the loci CAPN-316 and TG5. According to the obtained results, the periodic monitoring of the PYGM marker gene and of the allele Q204X the MSTN gene is propose; also it is important to implement strategies to confirm the usefulness of those markers associated with quality and productivity as a tool to complement breeding programs.
Keywords: Genetic defects; Marbling; Tenderness; DNA
Se determinaron las frecuencias alélicas y genotípicas de ocho marcadores localizados en los genes calpaína (CAPN, 4751 y 316), calpastatina (CASTT1) y tiroglobulina (TG5), asociados a calidad de carne, y en los genes, miostatina (MSTN, Q204X), arginino succinato sintasa (ASS), monofosfato sintasa (UMPS) y miofosforilasa (PYGM), asociados a enfermedades genéticas de ganado bovino. Se muestrearon 493 animales Charolais de registro de dos hatos ubicados en Sonora (n=157) y tres en Nuevo León (n=336). No se encontraron portadores de los alelos T-ASS y T-UMPS, pero sí portadores del alelo Q204X del gen MSTN en frecuencias de ≤ 1 % en las poblaciones de Sonora y de 8.6 a 14.4 % en las de Nuevo León. Además, se identificaron portadores del marcador del gen PYGM, en frecuencias del 6.5 y de 1.0 % para un hato de Sonora y otro de Nuevo León, respectivamente. El análisis de diferenciación génica pareado entre las poblaciones y con los cuatro loci mostró que hay diferencias altamente significativas dentro de poblaciones del noroeste (P<0.0001) y entre éstas y las del noreste (P<0.001), la cual es explicada principalmente por los loci CAPN-316 y TG5. De acuerdo a los resultados obtenidos se recomienda el monitoreo del marcador del gen PYGM y del alelo Q204X del gen MSTN, así como también implementar estrategias para confirmar la utilidad de los marcadores asociados a calidad y productividad como herramienta para complementar los programas de mejoramiento genético.
Palabras clave: Defectos genéticos; Marmoleo; Suavidad de la carne; ADN
Introduction
For cattle producers and all those interested in monitoring animal quality and genetic health, DNA testing is a widely available and important tool1,2. To date, DNA tests have been done identifying carriers of at least 40 genetic disorders in cattle1,3. In the Charolais breed, different cattle associations worldwide have reported the double muscle (DM) phenotype, a genetic condition produced by disruptive mutations in the myostatin gene (MSTN)4. Additional disorders reported for the Charolais breed include bovine citrullinemia (argininosuccinate synthetase - ASS); type-V glycogen storage disease (GSD-V); and uridine-5'-monophosphate synthase deficiency (UMSD). All these disorders are recessive and caused by mutations in specific genes5.
Among DNA tests currently used to predict meat quality and animal productivity, the most common are those focused to meat sensory features, such as tenderness and intermuscular fat content (marbling). The TG5 marker of the thyroglobulin gene is a C/T transition at the union consensus site of the RNA polymerase III, 537 bp upstream from the beginning of the thyroglobulin gene’s first exon (NW_001493192.1.g.290170C>T). This marker’s polymorphism is defined by alleles 2 and 3; allele 2 exhibits the GATC sequence while allele 3 has a GATT sequence (associated with greater marbling). Some studies have validated an association with marbling6, while others report no significant effect on this trait7. For tenderness, two markers localized in the micro-calpain 1 gene (CAPN1-316 and CAPN1-4751), and two others in the calpastatine gene (CAST) have received the most attention and have a validated effect on tenderness6. The marker CAPN1-316 is a G/C transversion at position 5709 of exon 9 in the CAPN1 gene, while CAPN1-4751 is a localized C/T transition at position 6545 between intron 17 and 18 of the same gene6. In both markers, the C allele has been reported and validated as favoring meat tenderness6. The marker UoG CAST is a G/C transversion at position 282 between exons 5 and 6, and the CAST-T1 marker is a localized G/A transition in the gene’s untranslatable 3' region. Allele C in UoG CAST, and allele A in CAST-T1 have been identified as favoring meat tenderness in cattle.
The present study objective was to typify registered Charolais cattle populations from Northwest and Northeast Mexico, using a SNPs panel associated with genetic diseases and meat sensory traits in order to determine their allele frequencies and understand their molecular genetic potential.
Materials and methods
Blood samples were taken from cattle at five registered Charolais breed ranches: two in the state of Sonora (SoR1 [n=83]; SoR2 [n=74]) in northwest Mexico, and three in the state of Nuevo Leon (NLR1 [n=79]; NLR2 [n=100]; NLR3 [n=157]) in the country’s northeast. Total sample size was 493 (21 bulls, 472 cows).
A Wizard® Genomic DNA Purification kit (Promega, Madison, WI, USA) was used to extract DNA from the blood samples. These were typified by allelic discrimination using the markers CAPN1-316 (G/C transversion), CAPN1-4751 (C/T transversion) and CAST-T1 (A/G transition), the allele Q204X (C/T transition), and primers and probes specific to each single nucleotide polymorphism (SNP) (Applied Biosystems, Mexico City, Mexico) (Table 1). Analysis was done using 250 ng DNA, 12.5 μl Taqman PCR master mix (Applied Biosystems), and 0.625 μl primer and probe mixture (Assay SNP mix). All assays were achieved in a thermocycler (ABI Prism 7000 Sequence Detection System) under the following conditions: 50 °C for 2 min; 95 °C for 10 min; and 40 two-step cycles of 92 °C for 15 s and 60 °C for 1 min. Genotype identification for each marker was done with the ABI Prism 7000 Sequence Detection System software.
Samples were also typified using four markers in PCR-RFLP assays. The specific primer sequences and restriction enzymes for SNPs detection were taken from the literature. The PCR runs for marker TG5 were done using the primers TG5U2 5’ggg gat gac tac gag tat gac tg 3' and TG5D1 5’gtg aaa atc ttg tgg agg ctg ta3', which generate a 545 bp fragment. The C/T transition was identified after digestion with the Mbo I enzyme. Allele 2 produces 17, 73, 177 and 278 bp fragments, while Allele 3 is characterized by a 73, 194 and 278 bp band pattern. For the GSD-V marker, the primers F-5'-CCA GGA AGA CCC TCA TTC CA-3' and R-5'-AGG GAA ACA CAC ACA CAG-3' were used5. The C/T transition in codon 489 of the myophosphorylase gene was detected with the Sty I enzyme. A band pattern of 119 and 133 bp was used to identify the TT homozygotic carriers, and a 252 bp pattern used for unaffected CCs. Markers associated with DUMPS were typified with the primers DUMPS-F 5'-GCA AAT GGC TGA AGA ACA TTC TG-3' and DUMPS_R 5'-GCT TCT AAC TGA ACT CCT CGA GT-3', while for ASS ASS-F 5'-GTG TTC ATT GAG GAC ATC-3' and ASS-R 5'-CCG TGA GAC ACA TAC TTG-3 were used5.
The C/T transition in the uridine-5'-monophosphate synthase associated with DUMPS was identified with the Ava I enzyme in which normal homozygotes exhibit a pattern of bands at 23, 36 and 19 bp, and recessive homozygotes have a pattern with 89 and 19 bp. For ASS, the C/T transition of codon 86 in the ASS gene protein was identified with the Ava II enzyme in which an indigested product indicated mutated carriers, and bands at 118 and 80 bp identified unaffected carriers5.
All amplifications were done in a 12.5 μl final volume containing 50 ng DNA, 2 mM MgCl2, 0.25 μM of each primer, and 0.125 U GoTaq DNA polymerase. A touchdown temperature profile was used in the assay: 95 °C for 10 h; 5 cycles at 95 °C for 45 min 65 °C for 45 min (decreasing 2 °C per cycle) and 72 °C for 45 min; 25 cycles at 95 °C for 45 min, 60 °C for 45 min and 72 °C for 45 min; and 72 °C for 10 h. Amplification was confirmed by electrophoresis in 1.5% agarose gel stained with SYBER® Gold; gels were viewed in a photodocumentor (Gel Logic 112, Kodak).
Enzymatic digestions were done using 10 μl PCR product and 2.5 U restriction enzyme specific to each polymorphism. Digestion band patterns were analyzed with electrophoresis in 4.5% Nusieve agarose gel prepared following manufacturer instructions (Karlan Research Products Co., Phoenix, AZ, USA).
Genotype and allele frequencies were then estimated with the Cervus 3.0 program9. Using the Genepop 4.2 program10, Hardy-Weinberg equilibrium (HWE) was calculated, and genetic differentiation between the studied populations was analyzed for the four meat quality-associated markers. In the genetic differentiation analysis, the tested null hypothesis was Ho= allele distribution identical across populations. For populations, the test was done automatically by pairs of populations using contingency tables. A non-biased estimate of the P value or a Fischer exact test was run10. Graphic illustration of differentiation by allele segregation among these markers was done with a correspondence analysis in the SAS ver. 9.0 program (SAS Institute Inc., Cary, NC, USA).
Results
Markers associated with genetic disorders
No carriers of ASS- or DUMPS-associated alleles were identified in the present data. However, some carriers were identified of genetic variants associated with GDS-V and DM (Table 2). Allele Q204X is associated with DM and was found in 28 (5.87 %) cows with frequencies in different herds varying from 0 to 14.4 %.
Table 2 Allele and genotype frequencies of four markers associated with genetic disorders and four associated with meat quality in registered Charolais cattle (%).
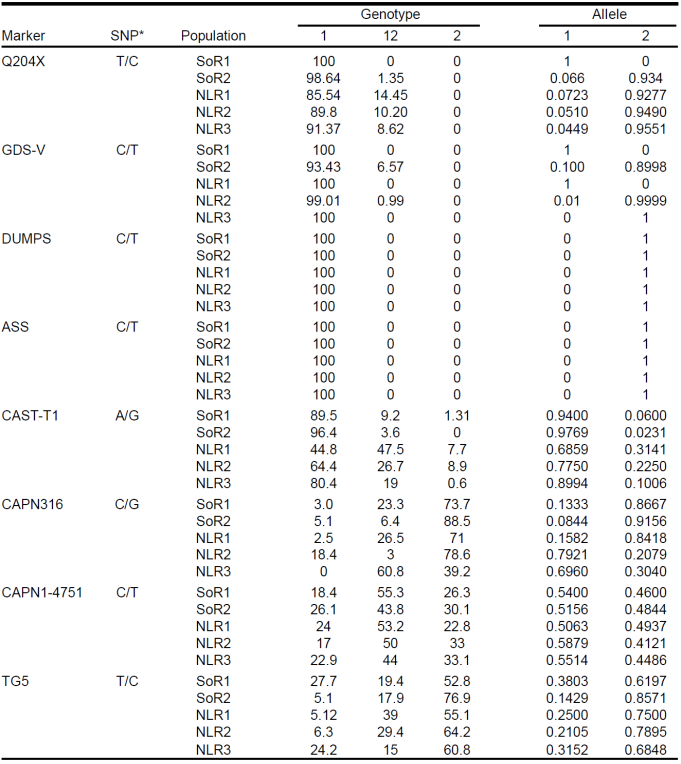
* Single nucleotide polymorphisms: ½.
When analyzed by region, the ranches in northeast Mexico had higher Q204X frequencies (8.6 to 14.4 %) than those in the northwest (maximum frequencies= 1 %). This discrepancy may be due to the source of the genetic material used in herd improvement. It is common in the northeast, particularly in the analyzed herds, to use European genetic material (mainly French semen). In some studies, carrier sire frequency is as high as 27 %11,12. To support this result, the pedigree data for the carrier cows identified in the study allowed them to be grouped into six families of half-sisters with French fathers. A biological sample was acquired from these French sires and DNA testing confirmed them to be Q204X carriers.
Five cows and one bull were also found to carry the GDS-V-associated marker (Figure 1). The highest frequency (6.49 %) was recorded in a herd in the northwest, with only one carrier in the northeast (1 %). Pedigree verification of two of the cows carrying this allele suggested it had been transmitted through the paternal line from a sire in the United States that was not included in this study.
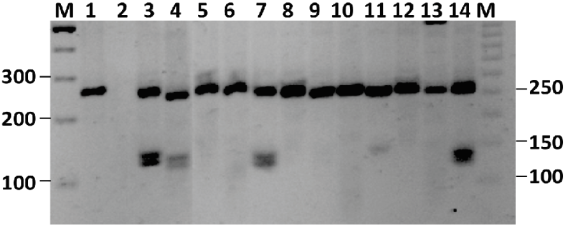
Rows 3, 4, 7 and 14 exhibit the 252, 133 and 119 bp pattern corresponding to carriers of the GDS-V associated allele in this population. Row 1= undigested PCR product; M= marker for 300, 200 and 100 bp molecular weights.
Figure 1 Identification of GDS-V carriers. PCR-RFLP results for individuals in SoR2 population.
Markers associated with meat quality
Paired genetic differentiation analysis between populations using the four analyzed loci showed highly significant differences within the two populations in the northwest (P<0.0001), and between these two populations and the three in the northeast (P<0.001) (Table 2). In the northeast, population NLR3 differed from the other two northeast populations (NLR1 and NLR2). Analysis by loci identified a difference between the four markers. The most significant was in the TG5 marker between SoR1 and SoR2, and between these two populations and NLR3. No differences between the populations were observed in marker 4751.
Allele segregation identified a strong influence from markers CAPN1-316 and CAST-T1 in differentiation between the northeast and northwest herds (Figure 2). It also indicated variability for the analyzed loci to be lowest among the two northwest herds.

Figure 2 Allele segregation of markers associated with meat quality in herds of Charolais cattle in northwest and northeast Mexico.
In the expected and observed heterozygosity data (Table 3), only two markers exhibited deviation in HWE. For TG5, this deviation was significant in the two northwest herds and NLR3, while for CAPN316 it was significant for SoR1, NLR2 and NLR3. All deviations were caused by a heterozygote deficit.
Discussion
Markers associated with genetic diseases
Cattle associations worldwide officially recognize genetic disorders in different cattle breeds and record them in animal registers. This is vital data that can be made available to animal and semen buyers, allowing them to exclude affected animals from breeding or adequately manage them within a herd. In this way, cattle associations can control dissemination of compromised genetic material and eventually eliminate carriers from the gene pool.
Frequency of the DM phenotype varies among different cattle breeds. This is because the traits associated to this phenotype have been selected for at varying intensities depending on the production goals of a given herd or population4. In the Charolais breed, the DM phenotype has been promoted mainly to produce carriers in terminal crosses12,13,14. Due to problems with dystocia, Charolais producers in Mexico have opted to reduce and even eliminate DM presence from their herds. Visual examination of animals is used to identify DM phenotype traits and manage them accordingly to each herd objective.
Allele frequencies in the present data ranged from 0 to 14 % and all carriers were heterozygotic. Presence of the allele in the analyzed Charolais populations could be due to the difficulty of identifying carrier animals from normal animals, because Charolais DM carriers do not exhibit the marked muscle development characteristic of the phenotype15. In these situations, DNA tests are a valuable tool because they generate data that can assist producers in making decisions based on herd objectives, taking into account the productive and reproductive advantages and disadvantages of the DM condition.
Carriers were also identified of the marker associated with GDS-V. This muscular disease is induced by a point mutation in the glycogen phosphorylase enzyme gene that causes intolerance to exercise, myalgia and recurrent myoglobinuria. Very little data is available on the prevalence of this recessive disorder in cattle, and most reports in Charolais animals are of carriers16. In a study of a carrier family in Charolais herds in New Zealand, presence of the allele was found to most probably be caused by import of animals from England and the United States17. The present study is the first report of carriers of the GDS-V-associated marker in Charolais cattle in Mexico. Of particular note is identification of a male carrier that currently has at least 38 registered progeny (34 males, 4 females) in the Asociación Charolais HerdBook de Mexico. To date, no DNA tests have been done to identify the recessive gene in these progeny. No reports exist on the productive consequences of GDS-V carriers, but their presence in herds is potentially damaging.
Markers associated with meat quality
Partially due to lack of completely defined criteria, data on meat quality in beef cattle in Mexico is limited18. However, the advent of biotechnology in the meat industry now allows application of molecular diagnosis of the genes associated with meat quality. This is an important first step in identifying the allele frequencies of these markers in registered cattle. The production process in Mexico requires that any genetic improvement program begin in the pure cattle breeds. For complex traits such as meat quality, improvement can be expected to appear in cattle for slaughter.
The meat quality markers analyzed here are included in commercial tests for meat tenderness and marbling, which is why the favorable effect of each allele variant has been reported6. Allelic and genotype frequencies observed in the present study, showed that the herds to have high to medium frequencies of the alleles reported as favorable for the markers CAST-T1 (allele A) and CAPN1-4751 (allele C). The allele reported as unfavorable for the markers CAPN316 and TG5 had the highest frequency (91.5 and 69.1 %, respectively).
The values in the present study are similar to those in a study of allele frequencies in French Charolais cattle for the markers CAST-T1 (high frequency) and CAPN316 (low frequency). The two markers of the CAPN1 gene have been reported to explain up to 25 % of genetic variation in meat tenderness19 and up to 18 % of phenotype variability6. However, other reports indicate that this effect depends on genetic background and the populations evaluated20.
Two of the four markers analyzed in the present study were included in a study of young Charolais bulls in Mexico21. The marker CAPN1-4751 had a significant effect on rib eye muscle area (REA) and intramuscular fat (IMF) measured by ultrasound, and the marker TG5 tended to associate with yield grade (YG). Research confirming this association, possibly supported by the National Council on Genetic Resources (Consejo Nacional de los Recursos Genéticos - CONARGEN), could help to measure meat quality traits using ultrasound.
The only study to date on the association of phenotypic variables to meat quality markers in Mexico supports a favorable association between markers 4751 and TG5, and the traits of cutting force and marbling7. However, this study’s limited sample size and is not considered as a validation study quantifying the effects of the genotypes of each marker in these traits. In the present study, the frequencies observed for the four studied meat quality markers justify implementation of management strategies aimed at increasing their frequencies, as well as design of studies to evaluate the effect of phenotype in marker-assisted management.
Although genetic differentiation was observed in the studied populations, and deviation was present in the HWE of two loci, it is unlikely that this can be explained by selection for meat quality traits (tenderness and marbling) since these are not included in genetic improvement strategies. The differentiation between the populations in the northwest (P<0.0001) and northeast (P<0.001) could be explained by the source of genetic material used in herd management. Producers in the northwest commented that they use Charolais animals and genetic material from the United States, with almost no influence from European genetic material. In contrast, producers in the northeast primarily use genetic material from France. This coincides with a study evaluating molecular genetic variation in three populations of Charolais cattle in Mexico22 in which genetic differentiation between populations was a consequence of selection of genetic material for herd management22.
All four quality markers used in the present study have been validated in the United States as predictors of meat marbling and quality. Commercial services are now available offering DNA tests for these and many other quality and productivity markers in cattle. This opens the possibility of producers or cattle associations employing these tests as tools to support genetic improvement programs, thus avoiding selection of individuals with unfavorable phenotypes. Further research is still needed on the association of these markers with traits in local populations, and validation of these associations to identify and quantify their effects on all traits of interest.
Conclusions and implications
No DUMP or ASS carriers were identified in the studied populations, showing them to have adequate genetic health. Some carriers of GDSV were found, highlighting the need for ongoing monitoring, especially of imported semen. Frequencies of the Q204X allele of the MSTN gene varied among herds, perhaps due to the source of genetic material used in herd management. Charolais producers need to understand the advantages and disadvantages of this allele in their herds, and use DNA testing to increase or eliminate it, depending on their production goals. Significant genetic differentiation between herds was present in the loci associated with quality and productivity. The frequencies of the alleles -reported and in some cases validated in the literature- identified in the studied populations and which are known to be favorable for different quality traits support implementation of strategies to confirm their use as a tool to complement genetic improvement programs in the Charolais breed in Mexico.
Acknowledgements
Thank to producers of the five studied Charolais herds and the Asociación Charolais HerdBook de México for providing study materials.
Financial support from FORDECYT 116152 and FOMIX Tamaulipas 177460.
REFERENCES
1. Ibeagha-Awemu EM, Kgwatalala P, Zhao X. A critical analysis of production-associated DNA polymorphisms in the genes of cattle, goat, sheep, and pig. Mamm Genome 2008;(19):591-617. [ Links ]
2. Strauss S. Genomics goes bovine. Nature Biotechnology 2010;28:6:540-543. [ Links ]
3. Ebegbulem VN, Ozung PO. Application of molecular markers in farm animal improvement: Prospects and challenges. Online J Anim Feed Res 2013;3(3):149-152. [ Links ]
4. Fiems LO. Double muscling in cattle: Genes, husbandry, carcasses and meat. Animal 2012;2:472-506. [ Links ]
5. Citek J, Rehout V, Hajkova J, Pavkova J. Monitoring of the genetic health of cattle in the Czech Republic. Vet Med Czech 2006;51(6):333-339. [ Links ]
6. Van Eenennaam AL, Li J, Thallman RM, Quaas RL, Dikeman ME, Gill CA, et al. Validation of comercial DNA test for quantitative beef quality traits. J Anim Sci 2007;(85):891-900. [ Links ]
7. Bonilla CA, Rubio MS, Sifuentes AM, Parra-Bracamonte GM, Arellano VW, Méndez MRD, et al. Association of CAPN1 316, CAPN1-4751 and TG5 markers with Mexican bovine meat quality traits. Genet Mol Res 2010;9(4):2395-2405. [ Links ]
8. Barendse W, Bunch R, Thomas M, Armitage S, Baud S, Donaldson N. The TG5 thyroglobulin gene test for marbling quantitative trait loci evaluated in feedlot cattle. Aust J Exp Agric 2004;(44):669-674. [ Links ]
9. Kalinowski ST, Taper ML, Marshall, TC. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol 2007;6:1099-1106. [ Links ]
10. Rousset, F. Genepop’007: a complete reimplementation of the Genepop software for Windows and Linux. Mol Ecol Resour 2008;8:103-106. [ Links ]
11. Allais S, Levéziel H, Payet-Duprat N, Hocquette JF, Lepetit J, Rousset S, et al. The two mutations, Q204X and nt821, of the myostatin gene affect carcass and meat quality in young heterozygous bulls of French beef breeds. J Anim Sci 2010;(88):446-454. [ Links ]
12. Parra-Bracamonte GM, Sifuentes-Rincón AM, Arellano-Vera W, Almanza-González A, De la Rosa-Reyna XF. Tipificación de tres marcadores genéticos de caracteres de importancia comercial en ganado Charolais: implicaciones en la ganadería para carne en México. Rev Colomb Cienc Pecu 2009;(22):257-266. [ Links ]
13. Phocas F. Genetic analysis of breeding traits in a Charolais cattle population segregating an inactive myostatin allele. J Anim Sci 2009;(87):1865-1871. [ Links ]
14. Dhuyvetter MJ, Frahm RR, Marshall MD. Comparision of Charolais and Limousin as terminal cross sire breeds. J Anim Sci 1985;(4):935-941. [ Links ]
15. Keele JW, Fahrenkrug SC. Optimun mating systems for the myostatin locus in cattle. J Anim Sci 2001;79:2016-2022. [ Links ]
16. Distl O. The use of molecular genetics in eliminating of inherited anomalies in cattle. Arch Tierz Dummerstorf 2005;48:(3):209-218. [ Links ]
17. Jolly RD, McSporran KD y Johnstone AC. Myophosphorylase deficiency (glycogen storage disease Type V) in Charolais cattle, N Z Vet J 2004;52(1):50-50. [ Links ]
18. Delgado EJ, Rubio MS, Iturbe FA, Méndez RD, Cassís L, Rosiles R. Composition and quality of Mexican and imported retail beef in Mexico. Meat Sci 2005;(69):465-471. [ Links ]
19. Allan MF, Smith TPL. Present and future applications of DNA technologies to improve beef production. Meat Sci 2008;(80):79-85. [ Links ]
20. Rousset LS, Denoyelle, Bernard-Capel C. Renand, Allais S, Journaux L, et al. Three French beef breeds effects of polymorphisms in the calpastatin and ì-calpain genes on meat tenderness. J Anim Sci 2011;89:1-11. [ Links ]
21. Muñoz-Mejía CY, Parra-Bracamonte GM, Sifuentes-Rincón AM, Martínez-González JC, López-Bustamante LA, Arellano-Vera W, et al. Indicadores genómicos y fenotípicos de calidad de la carne en bovinos Charolais de México. Rev Colomb Cienc Pecu 2012;(25):210-219. [ Links ]
22. Sifuentes-Rincón AM, Puentes-Montiel H, Parra-Bracamonte M. Assesment of genetic structure in Mexican Charolais herds using microsatellite markers. Electron J Biotechnol 2007;10:4-15. [ Links ]
Received: May 22, 2014; Accepted: July 09, 2014











 text in
text in 




