Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Revista mexicana de ciencias agrícolas
versión impresa ISSN 2007-0934
Rev. Mex. Cienc. Agríc vol.12 no.8 Texcoco nov./dic. 2021 Epub 02-Mayo-2022
https://doi.org/10.29312/remexca.v12i8.2684
Articles
Identification of F1 hybrids of poinsettia by RAPDS
1Facultad de Ciencias Agropecuarias-Universidad Autónoma del Estado de Morelos. Av. Universidad 1001, Cuernavaca, Morelos, México. CP. 62210.
2Escuela de Estudios Superiores de Xalostoc-Universidad Autónoma del Estado de Morelos. Av. Nicolás Bravo s/n, Parque Industrial Cuautla, Xalostoc, Ayala, Morelos, México. CP. 62717. Tel. 777 3297981.
Mexico is the center of origin of poinsettia Euphorbia pulcherrima Willd. ex Klotzsch, it is native to the state of Morelos. The molecular identification of hybrids determines the phylogenetic relationships between them. The objective of the present research was to identify F1 hybrids of E. pulcherrima using RAPD molecular markers. Thirty-nine F1 plants from the crosses of three varieties of sun poinsettias (Amanecer navideño, Belén and Juan Pablo) with three shade varieties (Festival red, Ice Punch and Burgundy) were used. Clustering analyses were performed on the relationship of matrices with the linkage method of unweighted pair groups with arithmetic mean (UPGMA). The OPA-07 primer is the one that allowed identifying the hybrid origin of 7 F1 plants of the Juan Pablo x Ice Punch cross. From the progeny of the crosses of Amanecer navideño x Ice Punch and Amanecer navideño x Festival red, the OPA primer identified 8 plants as hybrids. The RAPD primers used in the crosses obtained from Belén x Ice Punch, Belén x Burgundy and Belén x Festival red, failed to identify the parents with the hybrid progeny, the fragments did not show differences even between the same parents.
Keywords: diversity in F1 hybrids of poinsettia; F1 hybrids; identification; molecular markers; Poinsettias
México es el centro de origen de la nochebuena Euphorbia pulcherrima Willd. ex Klotzsch, es originaria del estado de Morelos. La identificación molecular de híbridos determina las relaciones filogenéticas entre ellos. El objetivo de la presente investigación fue identificar los híbridos F1 de E. pulcherrima mediante el uso de marcadores moleculares RAPD. Se utilizaron 39 plantas F1 provenientes de los cruzamientos de tres variedades de nochebuenas de sol (Amanecer navideño, Belén y Juan Pablo) con tres variedades de sombra (Festival red, Ice Punch y Burgundy). Los análisis de agrupamiento se realizaron sobre la relación de matrices con el método de ligamiento promedio aritmético de grupos de pares no ponderados (UPGMA) siglas en inglés. El iniciador OPA-07 es el que permitió identificar el origen híbrido de siete plantas F1 de la cruza Juan Pablo x Ice punch. De la progenie de las cruzas de Amanecer navideño x Ice punch y Amanecer navideño x Festival red, el iniciador OPA identificó a ocho plantas como híbridas. Los iniciadores RAPDs utilizados en las cruzas obtenidas de Belén x Ice punch, Belén x Burgundy y Belén x Festival red, no lograron identificar a los progenitores con la progenie híbrida, los fragmentos no mostraron diferencias incluso entre los mismos progenitores.
Palabras clave: diversidad en híbridos de nochebuena F1; híbridos F1; identificación; marcadores moleculares; nochebuenas
Introduction
Mexico is the center of origin of poinsettia (Euphorbia pulcherrima, Willd. ex Klotzsch), also known as Easter, Christmas Flower, Christmas Star or Poinsettia, it is native to the region that goes from the north of the state of Morelos to the municipality of Taxco, Guerrero (Martínez, 2001; Steinmann, 2002). In Mexico, poinsettia is a seasonal ornamental plant for the Christmas holidays, which contributes to the economic income of the country; annually, 25 million finished plants are produced, with a production value of 500 million pesos, it also generates direct and indirect jobs (Parra-Quijano et al., 2012).
This species is widely distributed throughout the tropical areas of Mexico (Lee, 2000), from sea level to 2 000 m of altitude, reaching its diversity and abundance in tropical deciduous forests; this species is distributed from the littoral of the Pacific Ocean to Guatemala, throughout the Gulf and center of Mexico (Steinmann, 2002). E. pulcherrima is a shrubby plant that reaches a height close to 5 m, has simple female flowers, without petals or sepals, surrounded by male flowers that are enclosed in a structure called cyathium, each flower forms a gland that attracts pollinators (Ecke et al., 2004; Taylor et al., 2011; Trejo et al., 2012).
Hybridization is one of the genetic improvement methods that takes advantage of the F1 generation from the cross between two parental populations P1 and P2 (Márquez, 1985). Usually, in a cross between two parents, genetic material is inherited from both; that is, the hybrid combines characters from both parents (Bai and Lindhout, 2007). Hybridization offers unique advantages in vegetatively propagated plants, in which recombination can be easily fixed (León, 2000). After hybridization, it is necessary to identify or detect similarities or differences of a cross (Watson and Eyzaguirre, 2002).
The traditional identification of plants is done by phenotypic characterization, a slow and limited procedure because the expression of quantitative characters is subject to strong environmental influence. Molecular markers allow identifying, classifying and taking advantage of the genetic diversity existing in plant genomes, since the differences or similarities at the DNA level between individuals are observed directly, as well as the expression influenced both by the dominant-recessive relationship and by the epistatic-pleiotropic interaction (Gutiérrez-Díez et al., 2009; Idrees and Irshad, 2014).
Nowadays, thanks to advances in molecular biology, molecular markers have become a tool for agricultural research in the genetic improvement of plants, such as genetic mapping, detecting mutant genes, improvement of crops assisted by markers, population history, epidemiology, food security, population studies, paternity tests and identification of cultivars, with this, the genetic variability of a plant material is classified and analyzed in order to take advantage of the diversity existing in plant genomes (Hartl and Jones 2005; Azofeifa-Delgado, 2006; Gutiérrez-Díez et al., 2009).
Zane et al. (2002) indicated that random amplification of polymorphic DNA (RAPD), simple sequences repeats (SSR), and amplified fragment length polymorphisms (AFLP) are fundamental tools for studying genetic variability. The study of genetic parameters such as molecular characterization can be performed for the identification of hybrids, to determine phylogenetic relationships and diversity at the genotype level and thus register new ornamental varieties with outstanding commercial desirable traits. Therefore, the objective of the present research was to identify F1 hybrids of E. pulcherrima using RAPD molecular markers.
Materials and methods
Plant material
In a previous study by Rodríguez et al. (2016), direct crosses were made, where the female parents were three sun poinsettias (Amanecer navideño, Belén and Juan Pablo) and the male parents were three shade poinsettias (Festival red, Ice Punch and Burgundy). From the progeny obtained, 39 F1 plants were selected as the object of study for their different and outstanding morphological characteristics from their male parents and female parent, such as bract color, bract shape and appearance of the plant. Samples of 200 mg of young leaves were taken from each of the 39 plants, leaves were also taken from the six parents, were weighed, labeled and frozen at -20 °C until DNA extraction.
DNA extraction and quality
For DNA extraction, the method described by Andrade et al. (2005) was used, which is based on CTAB. DNA integrity was estimated by 1% agarose gel (ultrapure GIBCO BRL) electrophoresis, the gel was stained with 1 (g ml-1 of ethidium bromide (Invitrogen®) and subsequently observed in an ultraviolet light transilluminator with a photodocumenter (Syngene® GVM20). The purity of the samples was determined by quantifying the amount of DNA with a NanoDrop 2000 spectrophotometer (Thermo scientific®). The DNA was diluted to a concentration of 20 ng µl-1 to be used in PCR reactions.
Fragment amplification
In a previous study Rodríguez et al. (2016), DNA amplification was performed with 68 random sequence primers of ten nucleotides from the OPA, OPB, OPC kit, plus 8 primers, of which, 10 were selected based on the quantity and brightness of the amplified RAPD fragments (OPA-07, 5’ GAA ACG GGT G 3’; OPA-11, 5’ CAA TCG CCG T 3’; OPA-12, 5’ TCG GCG ATA G 3’; OPB-07, 5’ GGT GAC GCA G 3’; OPB-08, 5’ GTC CAC ACG G 3’; OPB-09, 5’ TGG GGG ACT C 3’; OPB-11, 5’ GTA GAC CCG T 3’; OPB-20, 5’ GGA CCC TTA C 3’; OPC-06, 5’ GAA CGG ACT C 3’; OPC-13, 5’ AAG CCT CGT C 3’. The reaction components were as follows: 10 μl of dNTPs (5 μM of each dNTP, GIBCO BRL), 2.5 μl of PCR buffer (10x), 1.5 μl MgCl2 (3 mM), 2 μl primer (10 pmol μl-1), 0.3 μl (1.5 U) of native Taq DNA polymerase (Invitrogen), 4 μl DNA (20 ng μl-1) adjusting to 25 μl with 4.7 μl sterilized deionized water.
DNA amplification was carried out using RAPD markers, which are based on the polymerase chain reaction (PCR) technique. The separation of amplified fragments was done by 1.5% (w/v) agarose gel (Invitrogen®) electrophoresis, stained with ethidium bromide (10 μg l-1). Electrophoresis was performed at 75 V per 4 h with TAE 1X. The documentation of the gels was carried out in the same way as in the quality of the DNA.
Data analysis
The size of the DNA fragments that were generated by the RAPD primers was obtained using the Gene tools Labworks 4® program. The electrophoretic profiles generated by the parents and hybrids of each poinsettia variety were performed. The construction of the dendrogram was carried out to obtain the coefficient of similarity between progeny and parents, the data were analyzed based on the presence (indicated by 1) or the absence (indicated by 0) of amplified fragments with the selected decamer primers. Data were processed using the numerical taxonomy and multivariate analysis system (NTSYSpc 2.1). Clustering analyses were performed on the relationship of matrices with the linkage method of unweighted pair groups with arithmetic mean (UPGMA) (Avise, 1994).
Results and discussion
Identification of the progeny of Juan Pablo x Ice Punch and Juan Pablo x Festival red
With the primers used for DNA amplification of the crosses of Juan Pablo with Ice Punch and Festival red, monomorphic and polymorphic bands were obtained, 149 amplified fragments were obtained in total, of which 13% were polymorphic. The OPB-11 primer amplified the largest number of DNA bands with 23 amplified fragments and only 9% were polymorphic (Table 1), the primer that obtained the highest percentage of polymorphism was OPB-09, with 63%. The percentage of polymorphism ranged from 9 to 63%.
Table 1 Total, polymorphic DNA fragments and percentage of polymorphism for the identification of hybrid plants similar to the male parent of Juan Pablo x Ice Punch by RAPD.
| Primer | Amplified fragments | Polymorphic fragments | Polymorphism (%) | Marker fragments (pb)/plants identified per family |
| OPA-07 | 19 | 5 | 27 | IP (1971 bp/2, 3, 4, 7 and 8), IP (1391 pb/2, 3 and 4), IP (1101/1, 2, 3, 4 and 8), IP (1080/2) and IP (1 and 2) |
| OPA-11 | 18 | 3 | 17 | IP (2135 bp/2, 3 and 5) and IP (941 bp/3) |
| OPA-12 | 16 | 3 | 19 | IP (909 bp/ 6 and 7) |
| OPB-07 | 18 | 3 | 17 | IP (1924 bp/4) |
| OPB-08 | 8 | 3 | 38 | IP (749 bp/6) (666 bp/6 and 7) |
| OPB-09 | 8 | 5 | 63 | - |
| OPB-11 | 23 | 1 | 4 | IP (914 bp/3, 4 and 5) |
| OPB-20 | 14 | 3 | 21 | IP (1267 bp/1) |
| OPC-06 | 9 | 1 | 11 | IP (1979 bp/1, 2, 3 and 4) |
| OPC-13 | 16 | 2 | 13 | - |
The size of the bands that identified the hybrid origin of the F1 plants ranged from 446 bp to 2526 bp. The primers that allowed identifying the hybrid origin of the F1 plants of the Juan Pablo x Ice Punch cross were those shown in Table 2, being the OPA-07 primer the one that most identified plants from the cross, and the plants that could be identified as hybrids with other primers were the plants 2 JP x Ip, 3 JP x Ip, 4 JP x Ip, 5 JP x Ip, 6 JP x Ip, 7 JP x Ip and 8 JP x Ip. This condition of similarity of bands with the male parent allowed corroborating that hybridization between the female Juan Pablo parent with the male Ice Punch parent was obtained.
Table 2 Total, polymorphic DNA fragments and percentage of polymorphism for the identification of hybrid poinsettia plants of the crosses of the Belén parent by RAPD.
| Primer | Amplified fragments | Polymorphic fragments | Polymorphism (%) | Marker fragments (pb) |
| OPA-07 | 14 | 3 | 16 | - |
| OPA-11 | 16 | 4 | 25 | 2234 bp, 1894 bp, 1220 bp and 887 bp |
| OPA-12 | 9 | 2 | 22 | 1619 bp and 536 bp |
| OPB-07 | 10 | 1 | 10 | - |
| OPB-08 | 8 | 2 | 25 | 1035 bps |
| OPB-09 | 9 | 3 | 33 | 1382 bp and 1299 bp |
| OPB-11 | 7 | 2 | 28 | 1569 bp |
| OPB-20 | 13 | 3 | 23 | - |
| OPC-06 | 13 | 5 | 38 | - |
| OPC-13 | 14 | 5 | 36 | 609 bp |
The analysis of the conglomerate obtained (Figure 1) shows that, at a level of 0.26 of genetic distance, 6 groups form: 1) Juan Pablo and Festival red, 2) Ice Punch, 3) 2 JP x Ip, 4 JP x Ip, 3 JP x Ip, 6 JP x Ip, 7 JP x Ip, 8 JP x Ip,1 JP x Fr and 2 JP x Fr, 4) 3 JP x Fr, 5) 3 JP x Ip and 6) 1 JP x Ip. The group that concentrated more F1 plants was group 3. The hybrids most similar to each other (with a coefficient of 0.15) are 1 JP x Fr and 2 JP x Fr, these are the ones with the lowest genetic distance. In contrast, the plant that does not share genetic similarity with any parent or progeny is the male Ice Punch parent (Figure 1).
The above could explain why in the Juan Pablo x Festival red cross, less progeny was achieved (3), since by sharing greater genetic similarity, there is less possibility of fertilization due to genetic closeness. In this regard, Jiménez-Durán and Cruz-García (2011) mention that the strong genetic control exercised by sexual incompatibility systems greatly restricts introgression with parents and crosses with other members of the progeny.
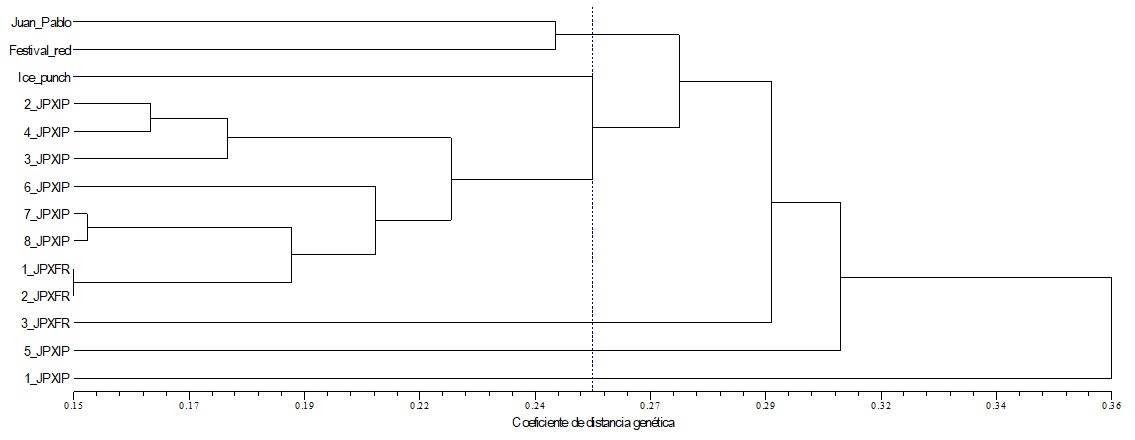
Figure 1 Relationship of the crosses generated on poinsettia, by Juan Pablo x Ice Punch (8 plants) and Juan Pablo x Festival red (3 plants) as determined by the UPGMA grouping analysis taking 149 RAPD fragments as a reference.
On the contrary, the female Ice Punch parent is in a quadrant separate from its progeny and the other parents, showing a longer distance of separation between them because Ice Punch is the product of an interspecific cross between other poinsettias and there is greater genetic distance. The distribution obtained in the crosses generated by Juan Pablo x Ice Punch and Juan Pablo x Festival red through a two-dimensional graphic distribution gave rise to four more detailed groups: 1) the male Ice Punch parent with the crosses of Juan Pablo x Ice Punch 2, 3, 4, 6, 7 and 8; 2) the female Juan Pablo parent with the cross 5 of Juan Pablo x Ice Punch; 3) the male Festival red parent with the crosses 1 and 2 of Juan Pablo x Festival red and 4) the crosses 1 of Juan Pablo x Ice Punch and 3 of Juan Pablo x Festival red (Figure 2).

Figure 2 Graphic distribution of two male parents Ice Punch and Festival red, one female Juan Pablo parent, 8 plants generated in the Juan Pablo x Ice Punch cross and 3 plants generated in the Juan Pablo x Festival red cross.
The RAPD markers failed to identify the progeny from the male Festival red parent, which may be due to the fact that hybridization between Juan Pablo and Festival red was not carried out, despite the fact that the hybridization process was taken care of so that they were not fertilized by other pollen from other plants or self-fertilized, there is a possibility that the plants are the product of self-fertilization and that is why they grouped together in group III, contrary to group I, where the male Ice Punch parent grouped with its progeny of Juan Pablo x Ice Punch, the uniformity of the behavior of this group and the identification with RAPD markers shows that hybridization was carried out, evidencing the genetic identity of the six progeny studied.
Identification of the progeny of Amanecer navideño x Ice Punch and Amanecer navideño x Festival red
The amplification of the RAPD primers used in the study of the crosses obtained from Amanecer navideño x Ice Punch and Amanecer navideño x Festival red revealed the presence of 146 fragments with a percentage of polymorphism of 34% (Table 3). The OPB-07 and OPB-20 primers were the ones that generated the greatest amplification of fragments (20) with polymorphism of 35% and 50% respectively, although the OPB-11 primer generated less fragments, it had 56% polymorphism, so the percentage of polymorphism varied from 27 to 56%.
Table 3 Total, polymorphic DNA fragments and percentage of polymorphism for the identification of hybrid poinsettia plants of the crosses of the parent Amanecer navideño by RAPD.
| Primer | Amplified fragments | Polymorphic fragments | Polymorphism (%) | Marker fragments (pb) |
| OPA-07 | 15 | 5 | 33 | IP (2179 bp/2 and 4), IP (1987 pb/2, 3, 4 and 7), IP (1531 bp/2, 3, 4 and 7), IP (855 bp/2, 4 and 5), |
| OPA-11 | 12 | 5 | 42 | IP (1507 bp/2, 3, 4, 6 and 7) |
| OPA-12 | 15 | 4 | 27 | IP (908 bp/5, 6 and 7), IP (815 bp/3, 4, 5, 6 and 7) |
| OPB-07 | 20 | 7 | 35 | IP (2455 bp/2, 3, 4, 5, 6 and 7), |
| OPB-08 | 6 | 2 | 33 | IP (661 bp/1, 4 and 7) |
| OPB-09 | 9 | 3 | 33 | IP (1270 bp/2, 4, 7) |
| OPB-11 | 19 | 5 | 56 | IP (1873 pb/ 2, 3, 5) |
| OPB-20 | 20 | 10 | 50 | Fr (812 bp/ 2, 3) |
| OPC-06 | 12 | 4 | 33 | - |
| OPC-13 | 18 | 5 | 28 | - |
The fragments that identified the progeny of the crosses of Amanecer navideño x Ice Punch and Amanecer navideño x Festival red ranged from 661 to 2455 bp. The primer that generated the largest number of fragments that identified hybrid plants was OPA-07, the percentage of polymorphic information of this primer was 33%. The primers that did not detect any hybrid obtained from the crosses were OPC-06 and OPC-13 (Table 3).
The plants that could be identified as hybrids with most of the primers were 2 An x Ip, 3 An x Ip, 4 An x Ip, 5 An x Ip, 6 An x Ip, 7 An x Ip, 2 An x Fr and 3 An x Fr. The above suggests that the crosses made of Amanecer navideño x Ice Punch were effectively achieved, since it was possible to identify its progeny with respect to the male Ice Punch parent, except for the plant 1 of this cross that did not show any fragment in common with the male Ice Punch parent.
The dendrogram generated (Figure 3) from the data obtained with the ten RAPD primers showed the genetic closeness between plant 5 An x Ip and 6 An x Ip, as well as plant 3 An x Ip and 4 An x Ip. Six groups formed at a genetic distance coefficient of 0.17: 1) Amanecer navideño with Ice Punch; 2) 3 An x Ip, 4 An x Ip, 7 An x Ip; 3) 2 An x Ip; 4) male Festival red parent, 1 An x Ip; 5) 2 An x Fr and 3 An x Fr; 6) 5 An x Ip, 6 An x Ip and 1 An x Fr.
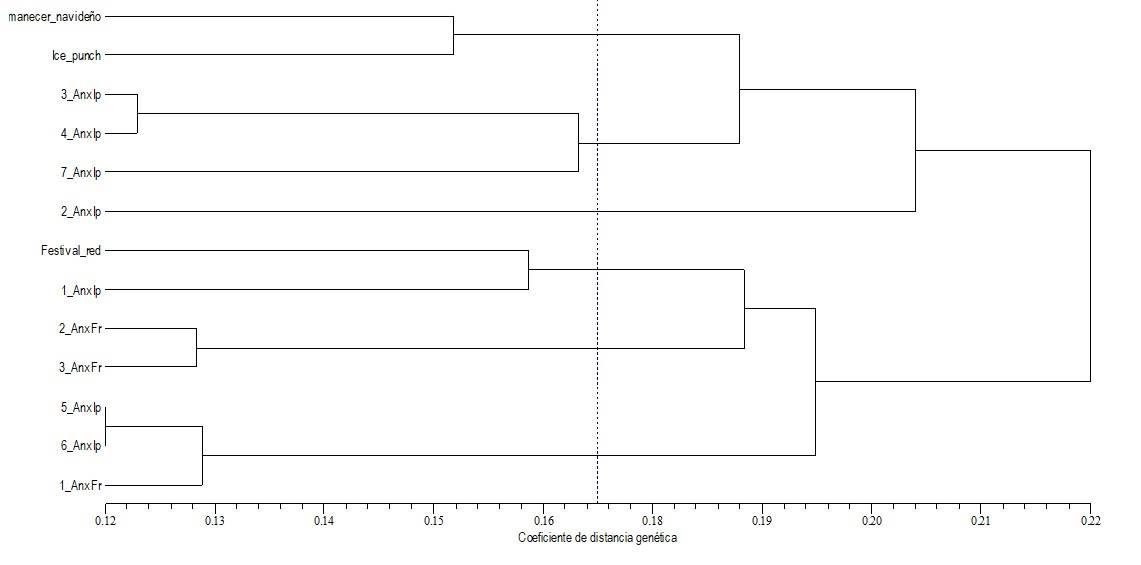
Figure 3 Relationship of the crosses generated on poinsettia, by Amanecer navideño x Ice Punch (7 plants) and Amanecer navideño x Festival red (3 plants) as determined by the UPGMA grouping analysis taking 146 RAPD fragments as a reference.
The plants most similar to each other (with a coefficient of 0.12) are 5 An x Ip and 6 An x Ip; as well as the progeny 3 An x Ip and 4 An x Ip, these are the ones that present the least genetic distance, in the same way, Amanecer navideño and Ice Punch parents share genetic similarity to a distance coefficient of 0.15. In contrast, the plant that does not share genetic similarity with its progeny or another parent is the male Festival red plant (Figure 3).
The conglomerate shows that the ten F1 plants of the crosses of Amanecer navideño x Ice Punch and Amanecer navideño x Festival red joined different groups, which shows genetic closeness between the three according to what was formed in the three groups formed: 1) the male Ice Punch parent; 2) the crosses of Amanecer navideño x Ice Punch 2, 3, 4 and 7; and 3 ) the female Amanecer navideño parent, the male Festival red parent with crosses 1, 5 and 6 Amanecer navideño x Ice Punch and the crosses 1, 2 and 3 of Amanecer navideño x Festival red (Figure 4).
Identification of the progeny of Belén x Ice Punch, Belén x Burgundy and Belén x Festival red
The amplification of the RAPD primers used in the study of the crosses obtained from Belén x Ice Punch, Belén x Burgundy and Belén x Festival red, revealed the presence of 113 fragments with a percentage of polymorphism of 26% (Table 2). The OPC-13 primer was the one that generated the highest amplification of fragments (14) with a polymorphism of 36%, but the primer that gave the greatest polymorphism was OPC-06 with 38% and 13 amplified fragments, the percentage of polymorphism varied from 10 to 38%.
The primers generated monomorphic marker fragments which were invariable in all the study plants, and these could not identify the parents with the hybrid progeny, the fragments did not show differences even between the same parents. The dendrogram generated (Figure 5) from the data obtained with the 10 RAPD primers showed the formation of 11 groups at a genetic distance coefficient of 0.18, the groups formed as follows: 1) the female parent Belén with Ice Punch; 2) the male Burgundy parent; 3) the male Festival red parent; 4) 2 Be x Bur; 5) 9 Be x Fr, 10 Be x Fr and 13 Be x Fr; 6) 11 Be x Fr; 7) 12 Be x Fr; 8) 1 Be x Ip, 1 Be x Bur, 4 Be x Fr, 5 Be x Fr, 2 Be x Fr, 3 Be x Fr; 9) 1 Be x Fr, 6 Be x Fr; 10) 7 Be x Fr, 8 Be x Fr; and 11) 3 Be x Bur.

Figure 5 Grouping of male parents (Ice Punch, Festival red and Burgundy) and the relationship of the crosses generated with Belén, as determined by the UPGMA grouping analysis taking 113 RAPD fragments as a reference.
The hybrid plants most similar to each other (with a coefficient of 0.09) were 1 Be x Bur and 4 Be x Bur, as well as 2 Be x Fr and 3 Be x Fr, Ice Punch and Belén parents share genetic similarity at a distance coefficient of 0.16 (Figure 5). The graphic distribution shows that the 18 F1 plants of the crosses of Belén x Ice Punch, Belén x Festival red and Belén x Burgundy are genetically different with respect to the four groups formed: 1) the male Ice Punch parent; 2) the hybrid 11 of Belén x Festival red; 3) the hybrids 1, 2, 3, 4, 5, 6, 7, 8 and 9 of Belén x Festival red and 1, 3, 4 of Belén x Burgundy; and 4) the crosses of the female Belén parent with the male Festival red parent and its crosses 1, 2, 3, 4, 5, 6, 7, 8 and 9; Belén x Ice Punch, and cross 1, the hybrids of Belén x Burgundy 1, 4 and 5 (Figure 6).
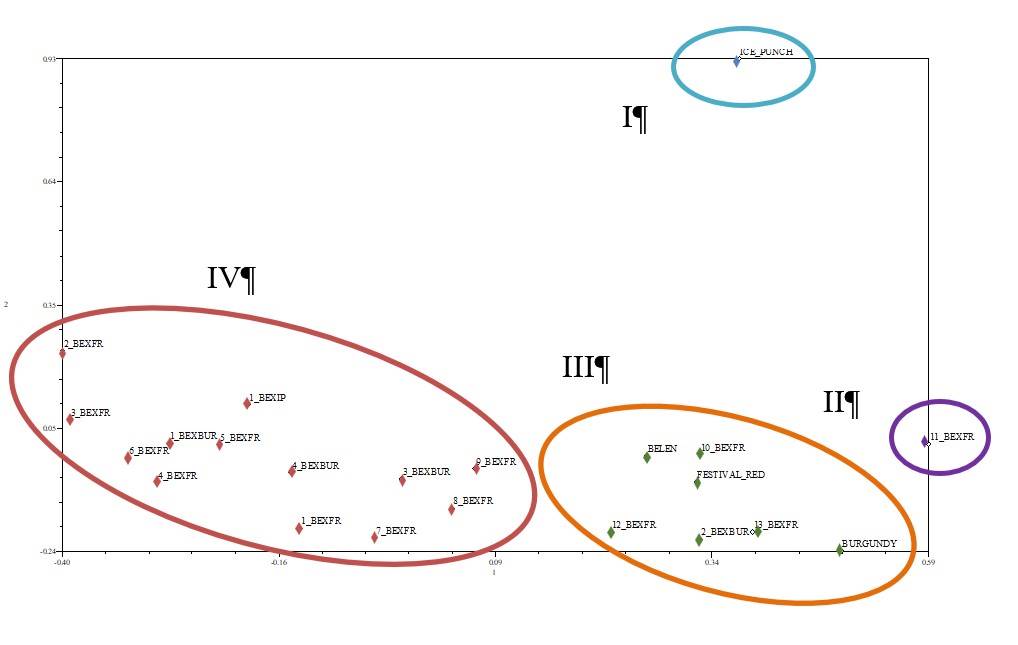
Figure 6 Graphic distribution of the male parent Belén x Ice Punch 1 plant generated, Belén x Festival red 3 plants generated, and Belén x Burgundy 14 plants generated in the crosses.
The primers selected for the parents Juan Pablo, Amanecer navideño and Belén and the plants originated from the poinsettia crosses of Juan Pablo x Ice Punch, Juan Pablo x Festival red, Amanecer navideño x Ice Punch, Amanecer navideño x Festival red, Belén x Ice Punch, Belén x Festival red and Belén x Burgundy, produced 408 RAPD fragments in total, of which 109 fragments were polymorphic with 27% polymorphism with 10 primers, these allowed identifying the F1 plants of Juan Pablo x Ice Punch, Juan Pablo x Festival red, Amanecer navideño x Ice Punch, Amanecer navideño x Festival red with 26 marker fragments, the crosses with Belén could not be identified with the 10 primers, since they generated marker fragments; however, these marker fragments did not allow identifying any hybrid, 11 marker fragments were produced, which could be evaluated with other primers.
This shows the discriminating capacity of RAPDs for the identification of poinsettia cultivars. The results of this research coincide with what was done by Jing-Tian et al. (1997), who identified poinsettia cultivars using RAPD markers, performed the amplification of 60 primers and obtained 69% polymorphism and conclude that their results indicate that RAPDs are efficient for the identification of Poinsettia cultivars and for the determination of genetic relationships between cultivars.
In another AFLP evaluation study in Poinsettia: polymorphism selection, analysis and identification of cultivars, they evaluated 81 poinsettia cultivars using 41 polymorphic AFPLs, of which they identified only 60 poinsettia cultivars, the other 21 were not identified (Parks and Moyer, 2004). In a study of comparison of RAPD and AFLP as methods of genetic identification of vines based on the study of fragments, they studied 57 varieties of vine with 80 primers, in their RAPD study, they mention that this result is remarkable since a correspondence between the genealogy and the information provided by the molecular analysis is verified and they conclude that both methods allowed differentiating all the cultivars analyzed (Narváez et al., 2000).
Conclusions
The molecular results obtained show a high level of identification of F1 hybrids by RAPDs, it was possible to determine the degree of similarity, thus demonstrating that RAPDs are an important tool for the identification of outstanding traits and differences in the morphological variation of F1 hybrids of poinsettia and it establishes the possibility of being able to obtain and register the breeder’s right of new varieties generated from a poinsettia genetic improvement program.
The perspective regarding the parents, both shade and sun varieties, is that the three sun varieties of poinsettia Juan Pablo, Amanecer navideño and Belén are a viable option as female parents for the search and formation of novel hybrids. While the shade varieties with which the greatest number of hybrids were obtained were Ice Punch and Festival red, but the hybrids between Belén and Festival red could not be identified with the RAPDs primers. In this context, it can be established that the poinsettia hybrids of Belén and Festival red are genetically similar and for the identification of the hybrids, it is convenient to apply and implement a subsequent study with the use of other primers or even other molecular markers for their identification.
Literatura citada
Andrade, R. M.; Villegas, Á.; Gutiérrez, A.; Carrillo, G. y García, A. 2005. Poliembrionía y marcadores raps para la identificación de plántulas cigóticas y nucelares en citrus. Agro Ciencia. 39(4):371-383. https://pdfs.semanticscholar.org/6d74/6c8bbc8ad2cee7dfff46c 8a964c794dcb981.pdf. [ Links ]
Avise, J. C. 1994. Molecular markers, natural history, and evolution. Chapman and hall. New York, N Y. 511 p. [ Links ]
Azofeifa-Delgado, A. 2006. Uso de marcadores moleculares en plantas; aplicaciones em frutales del trópico. Agron. Mesoam. 17(2):221-242. [ Links ]
Bai, Y. and Lindhout, P. 2007. Domestication and breeding of tomatoes: what have we gained and what can we gain in the future? Annal. Bot. 100:1085-1094. doi: 10.1093/aob/ mcm150. [ Links ]
Ecke, P.; Faust, J. E.; Higgins, A. and Williams, J. 2004. The Ecke Poinsettia manual.1st (Ed.). Ball publishing. Batavia, Illinois. 287 p. [ Links ]
Gutiérrez, D.; Martínez-Cerda, A. J.; García-Zambrano, E. A.; Iracheta-Donjuan, L.; Ocampo-Morales, J. D. y Cerda-Hurtado, I. M. 2009. Estudio de la diversidad genética del aguacate nativo en Nuevo León, México. Rev. Fitotec. Mex. 32(1):9-18. [ Links ]
Hartl, D. L. and Jones, E. W. 2005. DNA Structure and DNA manipulation. In genetics: analysis of genes and genomes. 5th (Ed). Sudbury: Jones and Bartlett Pub. 36-85 p. [ Links ]
Idrees, M. and Irshad, M. 2014. Molecular markers in plants for analysis of genetic diversity: a review. Eur. Academic Res. 2(1):1513-1540. [ Links ]
Jiménez-Durán, K. y Cruz-García, F. 2011. Incompatibilidad sexual, un mecanismo genético que evita la autofecundación y contribuye a la diversidad vegetal. Rev. Fitotec. Mex. 34(1):1-9. https://www.revistafitotecniamexicana.org/documentos/34-1/1r.pdf. [ Links ]
Jing-Tian, L.; Roger, S. and Nick, G. 1997. Identification of poinsettia cultivars using RAPD Markers. HortScience. 32(1):122-124 https://doi.org/10.21273/HORTSCI.32.1.122. [ Links ]
Lee, I. 2000. Phytoplasma casts a magic spell that turns the fair poinsettia into a Christmas showpiece. Online. Plant Health Progress. Doi: 1094/PHP-2000-0914-01-RV. [ Links ]
Márquez, S. F. 1985. Genotecnia vegetal, métodos, teoría y resultados. Tomo I. AAG. (Ed.). SA. México. 343 p. [ Links ]
Martínez, F. 2001. Manual práctico de la nochebuena. (Ed.). Oasis. 130 p. [ Links ]
Narváez, R. C.; Valenzuela, B. J.; Muñoz S. C. y Hinrichsen R. P. 2000. Comparación de RAPD y AFLP como métodos de identificación genética de vid basados en el estudio de fragmentos genómicos anónimos. Agric. Téc. 60(4):320-340. http://dx.doi.org/10.4067/S0365-28072000000400002. [ Links ]
Parks, J. E. and Moyer W. J. 2004. Evaluation of AFLP in poinsettia; polymorphismo selection, Analysis, and cultivar identification. J. Amer. Soc. Hort. 129(6):863-869. https://doi.org/10.21273/JASHS.129.6.0863. [ Links ]
Parra-Quijano, M.; Iriondo, M. and Torres, E. 2012. Ecogeographical land characterization maps as a tool for assessing plant adaptation and their implications in agrobiodiversity studies. Genetic resources and Crop Ev. 59(1):205-217. https://link.springer.com/content/pdf/ 10.1007/s10722-011-9676-7.pdf. [ Links ]
Rodríguez, R. T. J. M.; Andrade, R. O. G.; Villegas, T. I.; Alia, T. M. T.; Colinas, L. y Canul, J. K. 2016. Producción de frutos y calidad de semilla en cruzas de variedades de Euphorbia Pulcherrima Willd. Ex. Klotszch. Rev. Facult. Agron. (LUZ). 33:433-457. [ Links ]
Steinmann, V. W. 2002. Diversidad y endemismo de la familia Euphorbiaceae en México. Acta Bot. Mex. 61:61-93. https://www.redalyc.org/pdf/574/57406107.pdf. [ Links ]
Taylor, J. M.; Lopez, R. G.; Currey, C. J. and Janick, J. 2011. The poinsettia: history and transformation. Chronica Horticulturae. 51(3):23-28. https://www.actahort.org/chronica /pdf/ch5103.pdf#page=23. [ Links ]
Trejo, L.; Feria, T. P.; Olsen, K. M.; Eguiarte, L. E.; Arroyo, B.; Gruhn, J. and Olson, M. E. 2012. Poinsettia ‘s wild ancestor in the Mexican dry tropics: historical, genetic, and environmental evidence. Am. J. Bot. 99(7):1146-1157. https://doi.org/10.3732/ajb. 1200072. [ Links ]
Watson, J. W. and Eyzaguirre, P. B. 2002. Proceedings of the second international home gardens workshop: contribution of home gardens to in situ conservation of plant genetic resources in farming systems. Winzenhausen, Federal Republic of Germany. International plant genetic resources institute. Rome. 192 p. [ Links ]
Zane, L; Bargelloni, L. and Patarnello, T. 2002. Strategies of microsatellite isolation: a review. Mol. Ecol. 11:1-16. https://doi.org/10.1046/j.0962-1083.2001.01418.x. [ Links ]
Received: August 01, 2021; Accepted: November 01, 2021











 texto en
texto en 



