Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO
Share
Revista mexicana de ciencias agrícolas
Print version ISSN 2007-0934
Rev. Mex. Cienc. Agríc vol.10 n.2 Texcoco Feb./Mar. 2019
https://doi.org/10.29312/remexca.v10i2.1541
Article
In vitro activity to promote plant growth and biological control of rhizobacteria isolated from bermuda grass ruderal
1Departamento de Agricultura y Ganadería-Universidad de Sonora. Carretera a Bahía de Kino km 21, Hermosillo, Sonora, México. (merenteria@guayacan.uson.mx; andres.ochoa@unison.mx; miguel.barrera@guayacan.uson.mx; ernesto.fernandez@guayacan.uson.mx).
2Departamento de Investigaciones Científicas y Tecnológicas-Universidad de Sonora. Hermosillo, Sonora, México. (jmguzman@guayacan.uson.mx).
Currently, the use of plant growth promoting bacteria is a viable alternative to the indiscriminate use of agrochemicals. In order to obtain bacteria with the capacity to promote plant growth, in this work twelve strains with nitrogen fixation activity were isolated from the rhizosphere of bermuda grass (Cyonodon dactylon) ruderal. Based on the speed and intensity of the color change in the qualitative nitrogen fixation test, two isolates were selected for molecular identification and characterization. The amplification, sequencing and analysis of 16S rRNA ribosomal gene fragments allowed to identify Pseudomonas monteilii (Pm0710) and Chryseobacterium massiliae ca (Cm0711). Both isolates showed phosphate solubilization activity, production of indole-3-acetic acid, gibberellic acid and siderophores, alkaline phosphatase and ACC deaminase, as well as inhibition against: Fusarium brachygibbosum, F. falciforme, F. oxysporum and Ceratobasidium sp. Although Cm0711 was significantly higher than Pm0710 in most of the properties analyzed, the two bacteria have potential to be used as bio-inoculants; however, before validation must be done in vivo.
Keywords: Ceratobasidium; Chryseobacterium massiliae; Fusarium; Pseudomonas monteilii; PGPR
Actualmente, el uso de bacterias promotoras de crecimiento vegetal resulta una alternativa viable al uso indiscriminado de agroquímicos. Con el fin de obtener bacterias con capacidad de promoción del crecimiento vegetal, en este trabajo se aislaron doce cepas con actividad de fijación de nitrógeno a partir de la rizósfera de zacate bermuda (Cyonodon dactylon) ruderal. Con base en la rapidez e intensidad del cambio de color en la prueba cualitativa de fijación del nitrógeno, se seleccionaron dos aislados para su identificación molecular y caracterización. La amplificación, secuenciación y análisis de fragmentos del gen ribosomal 16S rRNA permitió identificar a Pseudomonas monteilii (Pm0710) y Chryseobacterium massiliae ca (Cm0711). Ambos aislados mostraron actividad de solubilización de fosfatos, de producción de ácido indol-3-acético, ácido giberélico y sideróforos, de fosfatasa alcalina y ACC deaminasa, así como de inhibición contra: Fusarium brachygibbosum, F. falciforme, F. oxysporum y Ceratobasidium sp. Aunque Cm0711 fue significativamente superior que Pm0710 en la mayoría de las propiedades analizadas, las dos bacterias tienen potencial para ser utilizadas como bioinoculantes; sin embargo, antes se debe realizar su validación in vivo.
Palabras clave: Ceratobasidium; Chryseobacterium massiliae; Fusarium; Pseudomonas monteilii; PGPR
Introduction
Bermuda grass (Cynodon dactylon) is planted in Mexico as fodder for cattle. This species was introduced into the American continent from Africa and due to its successful colonization of habitats, it is considered an invasive species (Van Devender et al., 2006). C. dactylon is able to grow in sodium soils of low fertility, because it forms interactions with some microorganisms in its rhizosphere, which favor its adaptation and promote its growth (Wang and Skipper, 2004; Singh et al., 2013).
The bacterial species present in the rhizosphere of plants, which grow in, on or around the root and which stimulate plant growth, are collectively known as plant growth promoting rhizobacteria (PGPR). PGPRs promote growth directly; through processes such as: nitrogen atmospheric fixation, phosphorus solubilization, mobilization of other essential minerals (for example, Zn and Fe) or regulation of plant hormone levels. In addition, they can exert an indirect effect due to their activity as agents of biological control against pathogens, predators and weeds (Glick, 2012).
One of the main challenges of the 21st century is sustainable agricultural production. The use of bio-inoculants is an environmentally friendly biotechnological option capable of reducing the use of agrochemicals. In this context, the objective of this work was to evaluate the activity of plant growth promotion and in vitro biological control of two bacterial species from the ruderal bermuda grass rhizosphere, through the analysis of nitrogen fixation capacity, solubilization of phosphates, production of indole-3-acetic acid, gibberellic acid, siderophores, enzymatic activity of alkaline phosphatase, ACC deaminase and confrontation with phytopathogenic fungi.
Material and methods
Isolation and selection of nitrogen-fixing bacteria
In October 2014, 10 healthy and vigorous Bermuda grass plants growing ruderally were selected and extracted in the Department of Agriculture and Livestock of the University of Sonora located at 29° 00’ 51.72” North latitude and 111° 08’ 01.10” West longitude, with an altitude of 149 m, in this site the climate is classified as BW (h’) hw (e’), which corresponds to the category of very arid, extreme and warm. The texture of the soil at the collection site is sandy loam. The aerial part of each plant was eliminated and approximately 1 kg of soil containing the roots was collected.
The soil was removed and the roots were washed with running water. The roots were cut into approximately 1 cm segments and rinsed 3 times for 10 min with sterile distilled water. Three root pieces were placed in screw-capped glass tubes, which contained 15 mL of semi-solid NFb medium (Döbereiner and Day, 1976) nitrogen-free. The tubes were closed, sealed with parafilm and incubated at 28 °C for 5 days. 1 mL of the microbial mass developed on the surface of the medium was collected and serial dilutions were seeded in Petri dishes containing solid NFb medium (agar, 15 g L-1).
After one week of incubation, the change from blue to green color around the colony was indicative that the isolate had nitrogen-fixing activity (Baldani et al., 2014). Twelve isolates with different morphologies were obtained and two of them, 0710 and 0711, were chosen to continue with phenotypic and biochemical characterization and molecular identification. The selection criteria were: greater intensity and speed in the change of color. The selected isolates were planted in Luria-Bertani medium (LB) for their conservation and subsequent analysis.
Phenotypic and biochemical characterization
Isolates 0710 and 0711 were cultured in total count agar for morphological analysis and characterized by the biochemical tests and carbon assimilation profiles of the API 20 E System commercial kit (BioMerieux Laboratory, France).
Molecular identification by sequencing a 16S rRNA gene fragment
The total DNA extraction of the selected isolates was carried out with the phenol-chloroform technique (Ausubel et al., 1994). The DNA was amplified by means of the polymerase chain reaction (PCR) using the F2C universal primers (5’-AGA GTT TGA TCA TGG CTC-3’) and C (5-ACG GGC GGT GTG TAC-3’), which amplify a fragment of around 1 600 bp. The reaction mixture contained: 20 ng of total DNA, 12.5 μL of GoTaq® Green Master Mix enzyme (Promega), 0.5 μM final concentration of each primer and water-free nucleases until a final volume of 25 μL was completed. The PCR conditions were: 1) initial denaturation at 95 °C for 2 min; 2) amplification, consisting of 30 cycles with the following steps: a) denaturation at 95 °C for 1 min; b) alignment at 55 °C for 1 min; and c) extension at 72 °C for 2 min and finally; and 3) final extension at 72 °C for 5 min. The BioRad C1000 thermocycler was used.
A second amplification was performed using the primers U1: 5’-CCA GCA GCC GCG GTA ATA CG-3’ and C, in order to obtain a smaller fragment (~ 900 bp), suitable for sequencing. As template DNA, 1 μL of the PCR products of the first amplification reaction was used. The amplification program was the same as the previous one. The final products were purified with a Wizard® SV Gel and PCR Clean-Up System kit (Promega Co.) and sequenced in both directions using again the U1 and C primers separately, with an ABI PRISM® 3730 Genetic Analyzer (Applied Biosystems). The sequences were compared with those of the NCBI GenBank database using the BLAST algorithm and aligned using the ClustalOmega program.
In vitro test of biological nitrogen fixation (FBN)
Isolates 0710 and 0711 were cultured in liquid enriched medium (1.5 g of meat peptone, 3 g of yeast extract, 6 g of gelatin peptone and 1 g of glucose in 1 000 mL of distilled-deionized water). A culture sample was taken and adjusted to a concentration between 105 and 106 CFU mL-1. 100 μL of the adjusted sample of each isolate was taken and inoculated in triplicate into 125 mL flasks with 50 mL of N-free thiamine-biotin medium. The flasks were incubated at 30 °C under agitation for 7 days.
The capacity of FBN was estimated according to Kanimozhi and Panneerselvam (2010), by determining the ammonia produced during the digestion process. 2 mL of the culture was placed in a Microkjeldahl flask, 2 mL of H2SO4 (d= 1.84), 1 mL of H2O2 (30%) and 1 g of digester mixture (100 g of Na2SO4 + 10 g of CuSO45H2O + 1 g of selenium) were added, at 350-375 °C and digested by boiling for 1.5 h. The digestion products were separated by distillation with 10 mL of NaOH 10 M and the ammonia produced was determined by titration with 0.0025 HCl M.
Qualitative phosphate solubilization assay
The phosphate solubilization test was carried out according to Malboobi et al. (2009). A needle tip was immersed in medium enriched with isolates 0710 and 0711 and was inoculated by stinging in the center of Petri dishes with the solid medium of Pikovskaya (15 g of agar, 10 g of glucose, 5 g of Ca3 (PO4)2, 0.5 g of yeast extract, 0.5 g of (NH4)2SO4, 0.2 g of KCl, 0.1 g of MgSO47H2O, 0.1 mg of MnSO42H2O, 0.1 mg of FeSO4 in 1000 mL of distilled water). Five boxes were planted per isolate and incubated at 30 °C for 7 days. The development of a clear zone around the colony was the sign of solubilization of inorganic phosphate.
Solubilization of inorganic phosphate (IPS)
Aliquots of 1 mL of cultures 0710 and 0711 were taken in liquid enriched medium and placed in 3 Erlenmeyer 500 mL flasks for each bacterium containing 200 mL of sterile Pikovskaya liquid medium. The flasks were kept under agitation (300 rpm) at 30 °C for 2 days. Sterile medium was used without inoculation as a negative control. After incubation, 10 mL of each flask was taken for centrifugation. The determination was carried out as described by Pote and Daniel (2000).
5 mL of each supernatant was taken and mixed with 800 μL of mixed reagent (50 mL of 2.5 M H2SO4, 15 mL of 4% ammonium molybdate, 30 mL of 0.1 M ascorbic acid and 5 mL of potassium tartrate solution 0.004 M) to quantify the release of phosphorus to the medium. A standard curve was prepared with known concentrations (0.15, 0.3, 0.6, 0.9, 1.2 and 1.5 μg mL-1), prepared from a stock solution containing 50 μg mL-1 of P. The absorbance of standard solutions was read and samples at 880 nm in a Spectronic 21D manual spectrophotometer (Milton Roy), using a reagent blank as a reference solution. The samples were proportionally diluted for reading in the graphic range.
Assay of the enzymatic activity of acid phosphatase (ACF) and alkaline (ALF)
The enzymatic activity of acid phosphatase and alkaline phosphatase was determined in triplicate using the modified test of Juma and Tabatabai (1988). A mixture of 1 mL of the supernatant obtained in the inorganic phosphate solubilization test of isolates 0710 and 0711 was made with 1 mL of 25 mM p-nitrophenyl phosphate (pNPP) and 4 mL of modified universal buffer [12.1 g of tris (hydroxymethyl) aminomethane, 11.6 g of maleic acid, 14 g citric acid and 6.3 g of H3BO3 in 488 ml of 1 M NaOH by 1 liter with distilled-deionized water. This buffer was adjusted to the desired pH and stored at 4 °C. Before use, it was diluted five times with distilled-deionized water]. The pH values of the buffer were 6.5 and 11, for the acid or alkaline phosphatase assay, respectively.
Each reaction mixture was incubated at 37 °C and after 1 h of incubation, 1 mL of CaCl2 0.5 M and 4 mL of NaOH 0.5 M were added to finish the reaction. The assay mixtures were filtered and their absorbance at 410 nm was recorded in a Spectronic 21D manual spectrophotometer (Milton Roy) to quantify the intensity of the yellow color due to the formation of p-nitrophenol (pNP). Quantification was performed using a pNP standard of known concentration and the results were expressed in units of activity (U), defining 1 U as the amount of enzyme required to release 1 μg of pNP in 1 mL of culture filtrate at the conditions of the assay (Prasanna et al., 2011).
ACC-deaminase assay
The activity of the ACC-deaminase enzyme was determined in triplicate according to the method developed by Penrose and Glick (2003). This assay determines the amount of (-ketobutyrate produced when the enzyme breaks down the 1-aminocyclopropane-1-carboxylic acid molecule (ACC). The number of μmoles of α-ketobutyrate produced in the reaction is determined by comparing the absorbance of a sample at 540 nm in a Spectronic 21D manual spectrophotometer (Milton Roy), against a standard α-ketobutyrate curve prepared in a range of 0.1 and 1 μmol.
Production of Indolaacetic acid (AIA)
It was carried out according to the method of Loper and Scroth (1986) and was carried out both in the absence and in the presence of 500 μg mL-1 of tryptophan. Previously bacterial isolates 0710 and 0711 were cultured in 3 mL of medium enriched for 24 h at 28 °C and 200 rpm. 20(L of that culture were transferred to 3 tubes with 5 mL of new enriched media for each isolate, without supplementation or supplemented with 500 μg mL-1 of tryptophan, incubated for 72 h at 28 °C and 200 rpm. After this time, the cultures were centrifuged for 10 minutes at 10 000 xg, 1 mL of supernatant was taken and mixed with 50 μL of orthophosphoric acid and 2 mL of the Salkowsky reagent (1 mL of 0.5 M FeCl3 in 50 mL of 35% HCIO4), incubating at 28 °C for 30 min.
At the end of that period, the appearance of a pink color indicated the presence of AIA. The quantification of AIA was performed by measuring the absorbance at 530 nm in a Spectronic 21D manual spectrophotometer (Milton Roy) with the help of a standard graph of AIA (Sigma) obtained in the range of 0 to 40 μg mL-1 (Ahmad et al., 2008).
Production of gibberellic acid (AG3)
Isolates 0710 and 0711 were cultured separately in tubes with 3 mL of enriched liquid medium, and incubated overnight at 28 °C and 200 rpm. 20 μL of each culture were transferred to 3 flasks of 50 mL of medium enriched per isolate and incubated under the same conditions for 72 h. The samples were centrifuged to remove the cell biomass. The determination was made according to Berrios et al. (2004). 1 mL of the supernatant was placed in a 10 mL volumetric flask and acidified by gauging with 3.75 M HCl. It was stirred vigorously for 10 s and allowed to react for 45 min to convert the gibberellic acid into giberelenic acid.
This compound was extracted once with 30 mL of ethyl acetate. The aqueous phase was discarded and the ethyl acetate phase was measured by a Milton Roy Spectronic 21D spectrophotometer (Milton Roy) at 254 nm. To estimate the amount of gibberellic acid produced, a calibration chart was prepared from standard solutions of: 50, 75, 100, 120, 150, 200, 240, 300, 400, 500 and 600 μg mL-1, prepared at from a stock solution of 100 mg AG3 in 250 mL of absolute ethanol. The solutions were acidified and extracted with ethyl acetate, similar to the sample. The values of absorbance against concentration were plotted and adjusted by linear regression.
Production of siderophores
For this test, cultures 0710 and 0711 were diluted in peptone water and planted at three equidistant points of three Petri dishes with agar-chromium azurol S (CAS) for each culture. The boxes were incubated at 28 °C for 48 h. After that time the siderophore producing bacteria, capable of fixing the iron, will produce an orange halo around the colonies in the blue medium (Schwyn and Neilands, 1988).
Production of hydrocyanic acid (HCN)
Bacteria 0710 and 0711 were cultured on glycine-modified King B agar medium (4.4 g L-1). A 8 cm diameter disc of Whatman filter no. 1 paper was placed inside the lid of the Petri box, previously soaked with a sterile solution of 1% picric acid in 10% sodium carbonate (filter paper and picric acid were sterilized separately). The plates were sealed with parafilm and incubated at 28 °C for 48 h. Three boxes were seeded by bacteria. The development of a reddish-brown color on the filter paper would indicate that the samples are positive for the production of HCN (Ahmad et al., 2008).
Antifungal activity assay
Isolates 0710 and 0711 were tested to determine the inhibitory effect of growth on the mycelial growth of Ceratobasidium sp., Fusarium oxysporum, F. falciforme and F. brachygibbosum, all of them previously isolated as causes of root rot in watermelon plants in Sonora, Mexico (Rentería-Martínez et al., 2018). The test was carried out on the complex agar medium (10 g of peptone, 40 g of D-glucose, 10 g of yeast extract, 18 g of agar in 1000 ml of water). The fungi were grown on potato dextrose agar (PDA). The pure bacterial cultures were cultured in liquid medium enriched at 28 °C for 24 h. 100 μL of each bacterial culture was spread on the surface of the complex agar.
Then, an 8 mm PDA disc covered with mycelium of each fungus was placed in the center of the Petri dish. At the same time, control experiments were performed, placing a disk of each fungus in the middle of the complex agar without bacteria. Three boxes were planted by bacteria. The diameter of each fungal colony was measured after an incubation period of 7 days at 28 °C. The inhibitory effect was calculated as the percentage of the inhibition rate: IR%= (CB)(100)/C, where C is the diameter of the fungal mycelium control and B the diameter of the mycelium grown in the presence of bacteria (Laslo et al., 2012).
Statistical analysis
The analysis of the results was carried out using the t test (for values of two independent variables), with a level of significance of 0.05%, using the IBM SPSS Statistics 24.0.0 software. Figure 1 referring to the inhibitory effect of bacterial isolates on pathogenic fungi was also performed analyzing the data with the same software.
Results and discussion
Microscopic analysis of isolate 0710 that grew in a nitrogen-free medium showed rod-shaped mobile cells typical of the genus Pseudomonas. The colonies in boxes with total count agar were: smooth, round and creamy yellow. The Gram stain was negative and the results of the biochemical tests of this isolate were: arginine dihydrolase (+), utilization of citrate (+), cytochrome oxidase (+) and oxidative metabolism of carbohydrates (-).
The sequences of the 16S rDNA gene fragments showed 100% homology with the registers: KM401858 and KF815702 of the GenBank, both accessions corresponding to Pseudomonas monteilii. Isolate 0711 showed 99% homology with the accessions KJ190166 and AF531766 of GenBank and was identified as Candidatus Chryseobacterium massiliae. Their colonies were yellow, with Gram negative cells, often in the form of paired rods, without motility and without spore formation. In biochemical tests, this species resulted in urease (+), gelatinase (+), production of indoleacetic (+) and fermentation of carbohydrates (-).
The isolate of P. monteilii was designated Pm0710 and that of Chryseobacterium massiliae ca. as: Cm0711. The results of the potential plant promotion characteristics of said isolates are summarized in Table 1. The two strains showed some properties of PGPR.
Table1. Characteristics of growth promotion of bacterial isolates plants.
| Characteristics | Pm0710 | Cm0711 |
| FBN | 52 ±10.6 a | 49 ±5.3 a |
| SFI | 143.9 ±2.2 b | 262.51 ±6 a |
| ACF | - | - |
| ALF | 21.2 ±1.8 b | 32.3 ±3.3 a |
| Siderophores | + | + |
| HCN | - | - |
| AIA (without tryptophan) | 2.4 ±0.1 b | 4.8 ±0.8 a |
| AIA (500 mg mL-1 of tryptophan) | 14.5 ±1.1 b | 44.4 ±8.6 a |
| AG3 | 40.9 ±3.17 a | 45.1 ±2.4 a |
| ACC | 640.7 ±37.4 b | 974.3 ±15.2 a |
Mean ± SD. Values with different letters in the same row are significantly different (p<0.05); FBN= biological nitrogen fixation in vitro (μg mL-1); SFI= solubilization of inorganic phosphate (μg mL-1); ACF= acid phosphatase activity (μg p-nitrophenol mL-1); ALF= alkaline phosphatase activity (μg p-nitrophenol mL-1); IAA= production of indoleacetic acid (μg mL-1); AG3= production of gibberellic acid (μg mL-1); ACC= 1-aminociclopropane-1-carboxylic acid deaminase activity (nmol α-ketobutyrate mg-1 protein h-1).
Penrose and Glick (2003) described that PGPR can facilitate the proliferation of their host plants through various mechanisms: 1) they fix atmospheric nitrogen and supply it to plants; 2) synthesize siderophores, which can provide iron to plants; 3) synthesize phytohormones (auxins, cytokinins and gibberellins); 4) solubilize minerals such as phosphorus and make them available for plants; and 5) synthesize enzymes capable of modulating the growth and development of plants.
In this work, Cm0711 and Pm0710 showed the capacity to fix atmospheric nitrogen during its in vitro culture (around 50 μg of N mL-1). According to Kim and Rees (1994), the enzymatic complex nitrogenase is responsible for the biological fixation of nitrogen (FBN). This complex consists of two metalloenzymes: dinitrogenase reductase, which provides electrons with high reducing power and dinitrogenase, which uses these electrons to reduce N2 to NH3.
The enzymatic system for N2 fixation varies between different bacterial genera. Three different systems have been identified according to their cofactor: a) Mo-nitrogenase; b) V-nitrogenase; and c) Fe-nitrogenase. The structural differences of these enzymes are the main responsible factors that control the efficiency of nitrogen fixation. However, since the Mo-nitrogenase system is found in all diazotrophic microorganisms, it can be said that it is the enzyme complex responsible for the greatest amount of biologically fixed atmospheric nitrogen.
The nitrogen fixed by free-living symbiotic diazotrophic microorganisms, associated or in the proximity of the roots, represents the most important contribution of nitrogen in natural ecosystems, being able to contribute up to 60 kg ha-1 year-1 of N to the soil. Although nitrogen-fixing bacteria associated in a symbiotic manner provide greater amounts of nitrogen required by the host plant in some terrestrial ecosystems (Glick, 2012), non-symbiotic nitrogen fixation may be the dominant form in the contribution in non-leguminous plants of zones arid (Cleveland et al., 1999). This is the case of C. dactylon in the study area.
There are two mechanisms for the solubilization of phosphorus by bacteria: mineral and enzymatic. The process of solubilization of inorganic phosphate (SFI) is associated with the release of organic acids of low molecular weight. The hydroxyl and carboxyl functional groups of these molecules chelate the cationic elements bound to the phosphate, ionizing and releasing the phosphate ions (Kim et al., 1998).
The enzymatic mechanism for the solubilization of phosphate groups linked to organic compounds involves the activity of two groups of enzymes: alkaline phosphatases (which act at basic pH) and acid phosphatases (which act at acidic pH). In this investigation, Cm0711 was significantly superior to Pm0710 in the capacity for inorganic phosphate solubilization (SFI) and in alkaline phosphatase activity. None of them presented acid phosphatase activity. In this regard, it is known that the roots of plants can obtain phosphorus from the soil due to their ability to release acid phosphatases and that they rarely produce appreciable amounts of alkaline phosphatases.
This fact suggests that the production of alkaline phosphatases represents a potential niche for the association of bacteria with the plant (Seema et al., 2013). Some bacteria associated with the roots of plants can produce 1-amino-cyclopropane-1-carboxylate (ACC) deaminase. This enzyme breaks the molecule by releasing ammonia and α-ketobutyrate. The ACC that is exuded by seeds, roots and leaves acts as a precursor of ethylene. Ethylene regulates many physiological processes associated with stress, and its presence at high concentrations can cause growth inhibition or death of the plant (Penrose and Glick, 2003; Glick, 2014).
Table 1 shows that both isolates had high levels of ACC deaminase activity, although Cm0711 was significantly higher than Pm0710. This suggests that both strains can decrease ethylene levels in plants during some stress event. The indoleacetic acid (AIA) secreted by the bacteria into the soil can modify the development processes regulated by the plant’s own auxins. The exogenous AIA alters the total content of bacterial AIA can increase the area and length of the root, providing the plant greater access to soil nutrients.
In contrast, rhizobacteria acquire nutrients that support their growth, as the growth of the root loosens the cell walls of plants, allowing them to exude a greater amount of nutrients (Patten and Glick, 2002, Glick, 2012). In this work, Cm0711 and Pm0710 showed the ability to synthesize AIA, increasing its production levels in the presence of tryptophan as a precursor. The production of AIA was significantly higher in Cm0711 than Pm0710.
Contrary to its effect as a growth promoter, AIA can induce the synthesis of ACC synthase, which catalyzes the formation of ACC in the plant. In this case, AIA would be acting as a promoter of ethylene synthesis. However, in the presence of PGPR, which synthesizes AIA and ACC deaminase, the ethylene levels in the plant may be lower than when the plants interact with PGPR that secrete AIA, but do not synthesize the deaminase ACC. The net result of this crosstalk between AIA and ACC deaminase, is that ACC deaminase facilitates the stimulation of growth by AIA effect, by reducing the levels of ethylene plants in the plant. This may result in greater adaptation to stress conditions (Glick, 2014).
In addition to the above, Cm0711 and Pm0710 showed the ability to synthesize gibberellins, observing levels around 40 μg mL-1 of gibberellic acid (AG3). Microbial gibberellins are typical secondary metabolites that can act as hormones, participating in the regulation of plant growth and development, including stem elongation, germination, dormancy, sexual expression and senescence of the fruits.
Both isolates also showed capacity to produce siderophores. Siderophores are small molecules secreted by numerous bacteria, actinomycetes and fungi. These compounds are specially designed to trap traces of ferric ions (Fe3+). Siderophores are secreted under conditions of iron limitation and form complexes with that element to be transported to the root cells. The iron is a little available element for the plants, since it forms very stable complexes in the ground. Several compounds (hydroxamates, phenolates, catecolates and carboxylates) can act as Fe3+ ligands.
One of the most studied mechanisms in the biological control of phytopathogens is the suppression of diseases mediated by siderophores. (Miethke and Marahiel, 2007; Angel et al., 2013). Another bacterial compound, which, due to its toxicity, in years ago was supposed to be a biological control agent is HCN. However, Rijavec and Lapanje (2016) demonstrated that the main contribution of HCN is in the sequestration of metals and phosphates, with the consequent indirect increase in the availability of nutrients, which is beneficial both for the rhizobacteria and their host plants. The property of synthesis of HCN does not exist in the two isolates evaluated.
Antifungal capacity
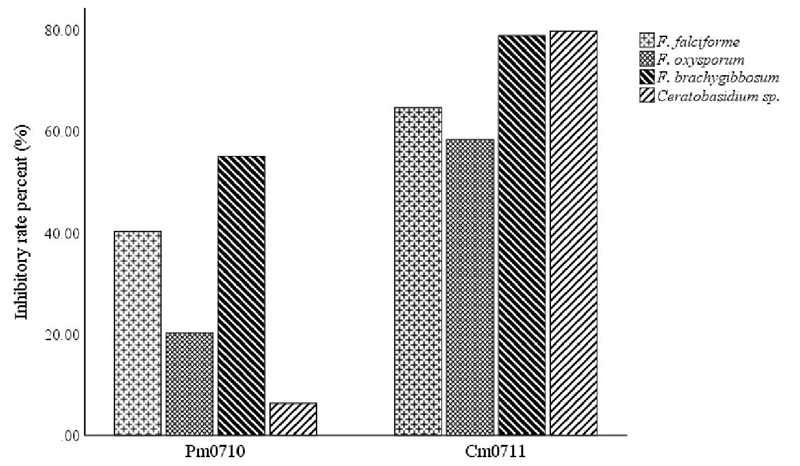
Cm0711 showed significantly higher levels of inhibition than Pm0710 in all tested pathogens. C. massiliae had inhibition rates of around 60 to 80% in all fungi. P. monteilii had an effect mainly on Fusarium brachygibbosum and F. solani (55% and 40%, respectively) and less on Ceratobasidium sp. and F. oxysporum, 6 and 20% respectively (Figure 1).
The biological control of phytopathogens is the main indirect mechanism for promoting the growth of plants by rhizobacteria. In general, competition for nutrients, exclusion of niches, induced systemic resistance and production of antifungal metabolites are the main modes of biological control activity of PGPR. Antifungal metabolites include antibiotics, enzymes that degrade the cell wall, chelating siderophores of Fe and HCN (Sindhu and Dadarwal, 2001; Ahmad et al., 2008; Ravindra et al., 2008; Bhattacharyya and Jha, 2012; Glick, 2012).
In our experiment, both Cm0711 and Pm0710 inhibited the growth of the four phytopathogens tested, however, the inhibitory effect of Cm0711 was significantly higher. The mechanisms by which these isolates exert control over plant pathogenic fungi remain unknown and should be elucidated in later work. Although it is known that some species of Chryseobacterium can act as PGPR (Shin et al., 2007; Dardanelli et al., 2010; Montero-Calasanz et al., 2013), there are no reports on the potential characteristics as a promoter of C. massiliae.
With respect to Pseudomonas monteilii, it has been observed that certain isolates can associate with rice plants and promote their growth due to their ability to fix nitrogen. (Habibi et al., 2014), while others significantly improve the length of the primary root, the outbreak and the number of lateral roots of the soybean sprouts (Wahyudi and Astuti, 2011). The phosphate solubilization capacity of P. monteilii strains has been previously reported (Ravindra et al., 2008; Dharni et al., 2014). Pandya and Desai (2014), found thirteen isolates capable of synthesizing gibberellic acid, in other works P. monteilii caused the inhibition of the growth of Sclerotium rolfsii by the production of diffusible antibiotics, volatile metabolites, hydrogen cyanide, siderophores and proteases (Rakh et al., 2011).
Another isolate produces 2,4-di-tert-butylphenol, an active volatile organic compound that acts as an inhibitor of β-tubulin in Fusarium oxysporum (Dharni et al., 2014).
Conclusions
The two species of bacteria isolated from the rhizosphere of Bermuda grass identified as C. massiliae and P. monteilii, showed characteristics of plant growth promotion that are evidence of their contribution to the adaptation and development of this plant. The ability to fix atmospheric nitrogen, the solubilization of phosphates, the production of phytohormones and the biological control of phytopathogens are desirable characteristics in microorganisms used in the formulation of bio-inoculants. However, before considering the use of Cm0711 and Pm0710 as PGPR, the evaluation of their behavior in vivo is required. The selection and use of native microorganisms capable of promoting the growth of plants is a necessary action to reduce the use of chemical fertilizers and to diminish some of the negative environmental impacts of conventional agriculture.
Literatura citada
Ahmad, F.; Ahmad, I. and Khan, M. S. 2008. Screening of free-living rhizospheric bacteria for their multiple plant growth promoting activities. Microbiol. Res. 163:173-181. https://doi.org/10.1016/j.micres.2006.04.001. [ Links ]
Ángel, J. M. R.; Reena, A.; Aysha, O. S.; Valli, S.; Nirmala, P. and Vinothkumar, P. 2013. Isolation of siderophore producing bacteria from rhizosphere soil and their antagonistic activity against selected fungal plant pathogens. Int. J. Curr. Microbiol. Appl. Sci. 2:59-65. https://pdfs.semanticscholar.org/8f88/199a5515686ab97854269c9dd9cff30d80c9.pdf. [ Links ]
Ausubel, F. M.; Brent, R.; Kingston, R. E.; Moore, D. D.; Seidman, J. G.; Smith, J. A. and Struhl, K. 1994. Curr. Prot. Mol. Biol. Wiley-Interscience, New York, USA. Section 2.4. [ Links ]
Baldani, J. I.; Massena, B. B. M.; Videira, S. S.; Boddey, L. H. and Divan, B. V. L. 2014. The art of isolating nitrogen-fixing bacteria from non-leguminous plants using N-free semi-solid media: a practical guide for microbiologists. Plant Soil. 384:413-431. https://link.springer.com/content/pdf/10.1007%2Fs11104-014-2186-6.pdf. [ Links ]
Berríos, J.; Illanes, A. and Aroca, G. 2004. Spectrophotometric method for determining gibberellic acid in fermentation broths. Biotechnology Letters. 26:67-70. https://link.springer.com/content/pdf/10.1023%2FB%3ABILE.0000009463.98203.8b.pdf. [ Links ]
Bhattacharyya, P. N. and Jha, D. K. 2012. Plant growth-promoting rhizobacteria. (PGPR): emergence in agriculture. World J. Microbiol. Biotech. 28:1327-1350. [ Links ]
Cleveland, C. C.; Townsend, A. R.; Schimel, D. S.; Fisher, H.; Howarth, R. W.; Hedin, L. O.; Perakis, S. S.; Latty, E. F.; Von Fischer, J. C.; Elseroad, A. and Wasson, M. F. 1999. Global patterns of terrestrial biological nitrogen (N2) fixation in natural ecosystems. Global Biogeochem. Cycles. 13:623-645. https://doi.org/10.1029/1999GB900014. [ Links ]
Dardanelli, M. S.; Manyani, H.; González-Barroso, S.; Rodríguez-Carvajal, M. A.; Gil-Serrano, A. M.; Espuny, M. R. and Ollero, F. J. 2010. Effect of the presence of the plant growth promoting rhizobacterium (PGPR) Chryseobacterium balustinum Aur9 and salt stress in the pattern of flavonoids exuded by soybean roots. Plant Soil. 328(1-2):483-493. https://link.springer.com/article/10.1007/s11104-009-0127-6. [ Links ]
Dharni, S.; Sanchita, A.; Maurya, A.; Samad, A.; Srivastava, S. K.; Sharma, A. and Patra, D. D. 2014. Purification, characterization, and in vitro activity of 2,4-Di-tert-butylphenol from Pseudomonas monteilii PsF84. Conformational and Molecular Docking Studies. J. Agric. Food Chem. 62:6138-6146. Doi: 10.1021/jf5001138. [ Links ]
Döbereiner, J. and Day, J. M. 1976. Associative symbiosis in tropical grasses: characterization of microorganisms and dinitrogen fixing sites. Proceedings of the First International Symposium on Nitrogen Fixation, Newton, W. E. and Nyman, C. J. (Eds.). Washington State University Press, Pullman. 518-536 pp. [ Links ]
Glick, B. R. 2012. Plant growth-promoting bacteria: mechanisms and applications. Scientifica. 1-15. http://dx.doi.org/10.6064/2012/963401. [ Links ]
Glick, B. R. 2014. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 169:30-39. https://doi.org/10.1016/j.micres.2013.09.009. [ Links ]
Habibi, S.; Djedidi, S.; Prongjunthuek, K.; Mortuza, M. F.; Ohtsu, N. O.; Sekimoto, H. and Yokoyoma, T. 2014. Physiological and genetic characterization of rice nitrogen fixer PGPR isolated from rhizosphere soils of different crops. Plant Soil. 379:51-66. http://dx.doi.org/10.1007/s11104-014-2035-7. [ Links ]
Juma, N. G. and Tabatabai, M. A. 1988. Phosphatase activity in corn and soybean roots: conditions for assay and effects of metals. Plant Soil. 107:39-47. https://link.springer.com/content/pdf/10.1007%2FBF02371542.pdf. [ Links ]
Kanimozhi, K. and Panneerselvam, A. 2010. Studies on isolation and nitrogen fixation ability of Azospirillum spp. isolated from Thanjavur district. Der Chem. Sinica. 1(3):138-145. http://www.imedpub.com/articles/studies-on-isolation-and-nitrogen-fixation-ability-of-azospirillum-sppisolated-from-thanjavur-district.pdf. [ Links ]
Kim, J. and Rees, D. C. 1994. Nitrogenase and biological nitrogen fixation. Biochemistry. 33:389-397. https://pubs.acs.org/doi/abs/10.1021/bi00168a001. [ Links ]
Kim, K. Y.; Jordan, D. and McDonald, G. A. 1998. Enterobacter agglomerans, phosphate solubilizing bacteria, and microbial activity in soil: effect of carbon sources. Soil Biol. Biochem. 30:995-1003. https://doi.org/10.1016/S0038-0717(98)00007-8. [ Links ]
Laslo, E.; György, E.; Gyöngyvér, M.; Tamás, E.; Ábrahám B. and Szabolcs, L. 2012. Screening of plant growth promoting rhizobacteria as potential microbial inoculants. Crop Prot. 40:43-48. https://doi.org/10.1016/j.cropro.2012.05.002. [ Links ]
Loper, J. E. and Scroth, M. N. 1986. Influence of bacterial sources on indole-3 acetic acid on root elongation of sugarbeet. Phytopathol. 76:386-389. https://www.apsnet.org/publications/phytopathology/backissues/documents/1986articles/phyto76n04-386.pdf. [ Links ]
Lorck, H. 1948. Production of hydrocyanic acid by bacteria. Physiol. Plant. 1:142-146. https://onlinelibrary.wiley.com/doi/abs/10.1111/j.1399-3054.1948.tb07118.x. [ Links ]
Malboobi, M. A.; Owlia, P.; Behbahani, M.; Sarokhani, E.; Moradi, E.; Yakhchali, B.; Deljou, A. and Heravi, K. M. 2009. Solubilization of organic and inorganic phosphates by three highly efficient soil bacterial isolates. World J. Microbiol. Biotech. 25:1471-1477. https://link.springer.com/article/10.1007/s11274-009-0037-z. [ Links ]
Miethke, M. and Marahiel, M. A. 2007. Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 71:413-451. doi:10.1128/MMBR.00012-07. [ Links ]
Montero-Calasanz, M. C.; Göker, M.; Rohde, M.; Spröer, C.; Schumann, P.; Busse, H. J.; Schmid, M.; Tindall, B. J.; Klenk, H. P. and Camacho, M. 2013. Chryseobacterium hispalense sp. nov., a plant-growth-promoting bacterium isolated from a rainwater pond in an olive plant nursery, and emended descriptions of Chryseobacterium defluvii, Chryseobacterium indologenes, Chryseobacterium wanjuense and Chryseobacterium gregarium. Int. J. Syst. Evol. Microbiol. 63(12):4386-4395. Doi 10.1099/ijs.0.052456-0. [ Links ]
Pandya, D. D. and Desai, P. V. 2014. Screening and characterization of GA3 producing Pseudomonas monteilii and its impact on plant growth promotion. Int. J. Curr. Microbiol. Appl. Sci. 3:110-115. https://www.ijcmas.com/vol-3-5/N.D.Pandya%20and%20P.V.%20Desai.pdf. [ Links ]
Patten, C. L. and Glick, B. R. 2002. Role of Pseudomonas putida indoleacetic acid in development of the host plant root system. Appl. Environ. Microbiol. 68:3795-3801. Doi:10.1128/AEM.68.8.3795-3801.2002. [ Links ]
Penrose, D. M. and Glick, B. R. 2003. Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol. Plant. 118:10-15. https://onlinelibrary.wiley.com/doi/pdf/10.1034/j.1399-3054.2003.00086.x. [ Links ]
Pote, D. H. and Daniel, T. C. 2000. Analyzing for dissolved reactive phosphorus in water samples: In: Pierzynski G. M. (Ed.). Methods of phosphorus analysis for soils, sediments, residuals, and waters. Southern Cooperative Series Bulletin No. 396. 91-93 pp. http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.454.4558&rep=rep1&type=pdf. [ Links ]
Prasanna, A.; Deepa, V.; Murthy, P. B.; Deecaraman, M.; Sridhar, R. and Dhandapani, P. 2011. Insoluble phosphate solubilization by bacterial strains isolated from rice rhizosphere soils from southern India. Int. J. Soil Sci. 6:134-141. Doi: 10.3923/ijss.2011.134.141. [ Links ]
Rakh R. R.; Raut, L. S.; Dalvi, S. M. and Manwar, A. V. 2011. Biological control of Sclerotium rolfsii, causing stem rot of groundnut by Pseudomonas cf. monteilii 9. Rec. Res. Sci. Tech. 3: 26-34. http://updatepublishing.com/journal/index.php/rrst/article/view/625. [ Links ]
Ravindra, N. P.; Raman, G.; Narayanan, K. B. and Sakthivel, N. 2008. Assessment of genetic and functional diversity of phosphate solubilizing fluorescent pseudomonads isolated from rhizospheric soil. BMC Microbiol. 8:1-14. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2625360/pdf/1471-2180-8-230.pdf. [ Links ]
Rentería-Martínez, M.E.; Guerra-Camacho, M, A.; Ochoa-Meza, A.; Moreno-Salazar, S. F.; Varela-Romero, A.; Gu tiérrez-Millán, L. E. and Meza-Moller, A. C. 2018. Multilocus phylogenetic analysis of fungal complex associated with root rot watermelon in Sonora, Mexico. Rev. Mex. Fitopatol. 36(2):233-255. Doi: 10.18781/R.MEX.FIT.1710-1. [ Links ]
Rijavec, T. and Lapanje, A. 2016. Hydrogen cyanide in the rhizosphere: not suppressing plant pathogens, but rather regulating availability of phosphate. Front Microbiol. 7(1785):1-14. doi:10.3389/fmicb.2016.01785. [ Links ]
Schwyn, B. and Neilands, J. B. 1988. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160:47-56. https://doi.org/10.1016/0003-2697(87)90612-9. [ Links ]
Seema, B. S.; Sayyed, R. Z.; Trivedi, M. H. and Gobi, T. A. 2013. Phosphate solubilizing microbes: sustainable approach for managing phosphorus deficiency in agricultural soils. SpringerPlus. 2:587. https://doi.org/10.1186/2193-1801-2-587. [ Links ]
Shin, D. S.; Park, M. S.; Jung, S.; Lee, M. S.; Lee, K. H.; Bae, K. S. and Kim, S. B. 2007. Plant growth-promoting potential of endophytic bacteria isolated from roots of coastal sand dune plants. J. Microbiol. Biotech. 17(8):1361-1368. http://www.jmb.or.kr/journal/viewJournal.html?year=2007&vol=17&num=8&page=1361. [ Links ]
Sindhu, S. S. and Dadarwal, K. R. 2001. Chitinolytic and cellulolytic Pseudomonas sp. antagonistic to fungal pathogens enhances nodulation by Mesorhizobium sp. Cicer in chickpea. Microbiol. Res. 156:353-358. https://doi.org/10.1078/0944-5013-00120. [ Links ]
Singh, K. V.; Pandey, C. and Singh R. P. 2013. Cynodon dactylon: An efficient perennial grass to revegetate sodic lands. Ecol. Eng. 54:32-38. https://doi.org/10.1016/j.ecoleng.2013.01.007. [ Links ]
Van Devender, T. R.; Felger, R. S.; Reina-Guerrero, A. L. and Sánchez-Escalante, J. 2006. Sonora: non-native and invasive plants. Invasive plants on the move. Controlling them in North America,.Proceedings of Weeds Across Borders. Van Devender, T. R.; Espinosa-García, F. J.; Harper-Lore, B. L. and Hubbard, T. (Eds.). Hermosillo, Sonora, México. 85-124 pp. [ Links ]
Wahyudi, A. T. and Astuti, R. I. 2011. Screening of Pseudomonas sp. isolated from rhizosphere of soybean plant as plant growth promoter and biocontrol agent. Am. J. Agric. Biol. Sci. 6:134-141. Doi: 10.3844/ajabssp.2011.134.141. [ Links ]
Wang, G. and Skipper, H. D. 2004. Identification of denitrifying rhizobacteria from bentgrass and bermudagrass golf greens. J. Appl. Microbiol. 97:827-837. Doi: 10.1111/j.1365-2672.2004.02368.x. [ Links ]
Received: January 2019; Accepted: March 2019











 text in
text in