Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Revista mexicana de ciencias agrícolas
versión impresa ISSN 2007-0934
Rev. Mex. Cienc. Agríc vol.9 no.3 Texcoco abr./may. 2018
https://doi.org/10.29312/remexca.v9i3.1212
Articles
Genetic diversity of maguey (Agave spp.) in the mountains and plains of northern Guanajuato
1Campo Experimental Bajío-INIFAP. Carretera Celaya-San Miguel de Allende km 6.5, Celaya, Guanajuato, México. CP. 38110. Tel. 01(800) 0882222, ext. 85269. (mandujano.andres@inifap.gob.mx; pons.joseluis@inifap.gob.mx).
3Técnico extensionista MasAgro-Guanajuato. Av. Irrigación 102-A, Col. Monte Camargo. Celaya, Guanajuato. México. CP. 38010. Tel. 01(461) 6226500, ext. 8290. (pul.gm4@gmail.com).
The agaves, endemic species of America with center of origin in Mexico are plants of semi-arid environments such as the mountains and plains of northern Guanajuato. In this subprovince are located the two municipalities of Guanajuato with designation of origin mezcal (DOM), which like tequila is distilled drink of agave with growing popularity inside and outside the country demanding more raw material and industrialization of processes including monoculture of species native or introduced agave, which reduces its genetic variability, putting at risk its diversity in the region. The objective of the research was to identify mezcalero agave species and their genetic diversity in the mountains and plains of northern Guanajuato, to generate base information to implement projects of dissemination, conservation, protection and rational use of agave, as well as to revalue this valuable resource, especially in the municipalities of San Felipe and San Luis of the Paz where there is a DOM. From 2014 to 2015, the field was identified, taxonomically characterized and collected samples of agave species distributed in the region. Later in the laboratory, they were analyzed using ISTR DNA markers, the Nei gene diversity was calculated. The taxonomic characterization allowed the identification of nine agave species and the diversity analysis determined that the agave populations of the mountains and plains of northern Guanajuato are poorly differentiated. 19% of the variation detected belongs to differences between species, the rest 81% represents genetic diversity within the species studied.
Key words: agave; genetic diversity; ISTR markers
Los agaves, especies endémicas de América con centro de origen en México son plantas de ambientes semiáridos como el de las Sierras y llanuras del norte de Guanajuato. En esta subprovincia se ubican los dos municipios de Guanajuato con denominación de origen mezcal (DOM), que igual que el tequila es bebida destilada de agave con creciente popularidad dentro y fuera del país demandando más materia prima e industrialización de procesos incluyendo el monocultivo de especies nativas o introducidas de agave, lo cual reduce su variabilidad genética, poniendo en riesgo su diversidad en la región. El objetivo de la investigación fue identificar las especies de agave mezcalero y su diversidad genética en las sierras y llanuras del norte de Guanajuato, para generar información base para implementar proyectos de divulgación, conservación, protección y aprovechamiento racional del agave, así como para revalorar este valioso recurso, sobre todo en los municipios de San Felipe y San Luis de la Paz donde se cuenta con DOM. De 2014 a 2015, en campo se identificaron, caracterizaron taxonómicamente y colectaron muestras de especies de agave distribuidas en la región. Posteriormente en laboratorio, se analizaron empleando marcadores ISTR de ADN, se calculó la diversidad génica de Nei. La caracterización taxonómica permitió identificar nueve especies de agave y el análisis de diversidad determinó que las poblaciones de agave de las sierras y llanuras del norte de Guanajuato están poco diferenciadas. 19% de la variación detectada pertenece a diferencias entre especies, el resto 81% representa diversidad genética dentro de las especies estudiadas.
Palabras clave: agave; diversidad genética; marcadores ISTR
Introduction
The genetic diversity of the species is the main reason for a given taxon to have the opportunity to evolve under changing environmental conditions at selection pressures (Eguiarte et al., 1999). The levels of genetic diversity or genetic variability of a population are defined by four main factors: dispersal or migration capacity, the reproduction and crossing mechanisms of the species, genetic drift and natural selection, of these, the mechanisms of reproduction and cross-linking, are those that to a greater extent determine the level of genetic variability in natural plant populations (Silvertown and Charlesworth, 2001), resulting in the great diversity of species.
Thanks to its enormous genetic variability, species and ecosystems, Mexico is considered as a megadiverse country and center of origin of diverse plant species, among which more than 75% of endemic species (approximately 150 species) figure agave, which is the most large genus of the Agavaceae family, originally from Mesoamerica (García-Mendoza, 2002), where these plants were one of the first to be harvested by their inhabitants (between 10 000-8 000 BC) as a source of food, drink, medicine, fuel, shelter, etc., promoting through human selection that Mexico become a center of domestication and diversification of agave (Aguirre et al., 2001; García, 2007).
The process of domestication of agave in Mexico gave rise to three groups of agave species: wild or maroon, semi-cultivated and cultivated (Parra and Tortolero, 2015), according to the above, in the state of Guanajuato different authors (Gentry, 1982; Parra, 2002; Parra, 2004; Hernández et al., 2012; García, 2014; Gámez, 2014) have reported the presence of 22 species of the agave genus, of which, 25% belong to the group of wild agaves that naturally distributed mainly in the semi-arid region of northern Guanajuato including San Felipe and San Luis of the Paz municipalities with denomination of origin mezcal (DOM) and that as in the Potosino-Zacatecano plateau have evolved as native vegetation and its use it has been through the collection, being the case of the Agave Salmiana subsp. Crassispina, Agave scabra potosiensis and Agave lechuguilla.
In the north of Guanajuato, wild agaves coexist with semicultivated species of agave that have been brought from other areas of the country and have been established in soil and water conservation works, and as in the rest of the state, have been established as living fences around the properties of those who take advantage of them for their mead, pulque, fodder, fibers, etc., in this group is Agave salmiana subsp. salmiana, the Agave mapisaga spp. mapisaga and the American Agave subsp. American, Agave inaequidens, among others. In Guanajuato, the most important Agave cultivated species is Agave tequilana weber Var. Blue, which is a species belonging to 75% of the introduced species, whose distribution occurs in the southwest of the state, in the municipalities with denomination of origin tequila (DOT).
Tequila and mezcal are distilled agave drinks, symbols of Mexicanness, which in recent years have gained popularity inside and outside the country, demanding a greater supply of raw materials to support the increase in their production (Eguiarte and González, 2007), which should be based on sustainable schemes that reduce social, economic and environmental impacts. An irrational exploitation of agave can generate problems of environmental degradation such as erosion, fragmentation of habitat and loss of genetic diversity (Bellon et al., 2009).
The reduction of the genetic variability of the agaves and in general of the species generates susceptibility to attacks of pests and diseases as well as little flexibility before climatic changes that increase the risk of their extinction (Erazo and Cárdenas, 2013). The knowledge and understanding of the genetic variability of the species of any region is vital to: determine its richness and distribution, plan harvesting and conservation strategies, as well as to diminish the negative effects caused by the proliferation of pathogens, invasive species and the introduction of improved and genetically modified varieties on populations of native plants and animals (Piñero et al., 2008).
In this sense, molecular markers are powerful biotechnological tools for the knowledge of biological diversity (Espinel and Aragones, 1997). These can be defined as specific sequences of deoxyribonucleic acid (DNA) with a specific location (locus) in a chromosome, whose genetic inheritance or effect can be quantifiable or observable (Dreisigacker, 2013). There are two types of molecular markers biochemists (proteins and izoenzymes) and DNA markers, the latter are based on the analysis of differences or similarities in small DNA sequences between individuals and have advantages such as: they work directly with the genetic base of the variations, they have a high degree of polymorphism, they are phenotypically neutral, they are not affected by the growth stage of the individuals, they are independent at the time of the year in which the analysis is carried out, they are a non-destructive technique due to the small amount of material they use, their cost is relatively low and they are accessible (Godoy, 2009).
The techniques used by molecular DNA markers are diverse and give rise to the name of the different types that exist, some examples of them are: polymorphism of the length of the restriction fragments (RFLP), random amplification of the polymorphic DNA (RAPD), polymorphism in the length of the amplified fragments (AFLP), microsatellites or simple repeated sequences (SSR), random amplification of the microsatellite polymorphism (RAMPO), repeated and marked reverse sequences (ISTR), etc (Rallo et al., 2002). The ISTRs are dominant markers based on retrotransposons, which are sequences that auto-replicate and reinsert in different parts of the genome via ribonucleic acid (RNA), play an important role in the mutation, evolution and organization of living beings (Torres et al., 2006).
In comparison with other markers, ISTRs have advantages such as their application to a broad spectrum of organisms, high reproducibility, use universal primers, do not require high quality or quantity of DNA, amplify a large number of loci (5-100), it is not necessary radioactivity for visualization and detect a considerable polymorphism (Demey et al., 2004; Torres, 2009).
Some of these molecular markers have been used to study the genetic diversity of agave, for example, Cuevas and Flores (2006) used the AFLP technique to compare different species and varieties of agave with Agave tequilana Weber Var. Blue found that this species shares close genetic relationships with Agave angustifolia Haw and large differences with species and varieties of A. americana L., A. sisalana Perrine ex Engelm, A. salmiana Otto et Salm., A. potatorum Zucc. and A. karwinskii Zucc. This same technique was used by Barraza et al. (2006) to analyze the genetic variability of populations of Agave angustifolia Haw. from the Sonoran Mountains of Mexico, finding high levels of variability.
On the other hand, Torres (2009) using the ISTR technique characterized populations of natural distribution and a lot of nursery seedlings of Agave durangensis to determine their genetic variability, finding the existence of great genetic diversity within said populations and seedlings, suggesting to Agave durangensis as an agave complex and not as a species. Infante et al. (2006) compared the results of genetic variability of different species of agave using two different techniques: AFLP and ISTR, finding a correlation of 0.9018 between the distances generated with AFLP and ISTR, which shows that both types of markers possess high levels of reliability for be used as a method to study the genetic variability between different or the same species of the agave genus.
The objective of this research was to identify the mezcalero agave species and determine their genetic variability in areas of high and good potential aptitude for the development of agave mezcal species (Gámez et al., 2014) in the physiographic subprovince of mountains and plains of the North of Guanajuato.
Materials and methods
The mountains and plains of northern Guanajuato is a physiographic subprovince of the Central Mexican Table that occupies about 38% of the territory (13 001 km2) of the state of Guanajuato covering 20 municipalities of the central-northern region of the state, extending in part of the states of San Luis Potosí and Querétaro. This subprovince is characterized by its semi-arid climate, landscapes dominated by steep mountain ranges, ravines, gorges and plateaus of volcanic origin, with elevations between 1 000 and 3 300 masl, as well as xerophilous scrub vegetation composed mainly of mesquites, huizaches, nopales and magueys.
From 2014 to 2015, trips were made to wild populations, magueyeras established for the purpose of reforestation and soil conservation, magueyeras located on margins of roads or borderias of cultivation plots, as well as in commercial agave plantations located on the surface classified as high and good potential aptitude for the development of agave mezcal species (Gámez et al., 2014). Each sampling site was georeferenced with a geopositioning (Garmin-20®).
Using the keys of Gentry (1982) as well as visual references of the digital agave collections of Julia Etter and Martin Kristen, the different species of agave found in each site were identified taxonomically and a sample of vegetal material was collected, which consisted of a piece of 100 g of the basal part of a leaf. The samples were preserved on ice until they were frozen at -80 °C in a REVCO® ultra-freezer for further processing in the laboratory of Molecular Markers of the Bajío Experimental Field of the INIFAP.
The CTAB protocol (hexadecyltrimethylammonium bromide) (Doyle and Doyle, 1987) modified with STE (sodium chloride-tris-EDTA) was used (Falcón and Valera, 2007), to which three modifications were also made: 1) eliminate twice the supernatant; 2) maintain the eppendorf tube for 15 min after adding RNAase and proteinase; and 3) dilute the DNA in TE-1X, in a solution of salts at low molarity. The quality of the DNA extractions was checked using a Nano Drop 8000® spectrophotometer using as parameters a purity of 1.8 to 2.1 μg ml-1 at 260/280 nm wavelength and a concentration greater than 50 μg ml-1, which subsequently Dilution was homogenized at 15 μg ml-1.
The quality of the dilutions was checked by electrophoresis of 4μl of genomic DNA in combination with 3μl of Biotina® as loading buffer and a lambda λ marker at a concentration of 25μg ml-1 in 1.5% agarose gels for 35 min at a constant power of 130V in the Power Pac 3000® unit using TBE 1X run buffer. The resulting gels were stained with ethidium bromide and photographed with UV rays with the Bio Rad® transilluminator.
The sequences and combinations of the ISTR markers used to determine the genetic variability of the mezcalero agaves in the mountains and plains of northern Guanajuato, were the same as those used by Torres (2009) in the complex of Agave duranguensis and Montalvo et al. (2010) in seedlings of Pilosocereus sp. These sequences are presented in Table 1.
Table 1 Sequence of the ISTR markers used by Torres (2009) and Montalvo et al. (2010).
| Initiator (Torres, 2009) | Initiator (Montalvo et al., 2010) | |||
| Sequence | Nom. | Sequence | Nom. | |
| d5’ (TTACCTCCTCCA TCT CGT AG) 3’ | F9 | d5’ (GTC GAC ATG CCA TCT TTC) 3’ | 3 | |
| d5’ (TAA GCA AGC ATC TCG GAG) 3’ | F10 | d5’ (TAT AGT ACC TAT TGG GTG) 3’ | F4 | |
| d5’ (ATC AGC AAG GTC TGT AAA GC) 3’ | B1 | d5’ (ATA TAT GGA CTT AAG CAA GC) 3’ | F5 | |
| d5’ (GGT TCC ACT TGG TCC TTA G) 3’ | B6 I | d5’ (GTA TTG TAC GTG GAT GAC ATC) 3’ | F6 | |
| d5’ (ATA CCT TTC AGG GGG ATG) 3’ | B8 | d5’ (ATT CCC ATC TGC ACC ATT) 3’ | B3 | |
| d5’ (GCA CTC CAC CAA GAA TAC C) 3’ | F1 | d5’ (ATA TAT GGA CTT AAG CAA GAC) 3’ | B6 II | |
F= forward; B= reverse.
The combinations of the initiators were as follows: F9/B6I, F10/B1, F1/B6I, F10/ B6I, F10/B8 (Torres, 2009) and F3/B3, F4/B3, F5/B3, F6/B3, F6/B6I (Montalvo et al., 2010). In addition, combinations formed using initiators of both authors were tested: F6/B8, F6/B6II, F5/B8, F5/ B6ll, testing a total of 14 combinations.
To find the optimal temperature of alignment (TM), the 14 combinations of primers in the DNA of the species Agave salmiana and Agave americana were tested using alignment temperatures: 40, 45, 50, 55, 60 and 65 °C, in 40 cycles.
For the amplification of the ISTRs through the polymerase chain reaction (PCR) a mixing volume of 25 μl composed of 10.8 μl of sterile distilled water, 2.5 μl of 10X buffer, 2 μl of DNTP’S was used. 2.5 μM, 1.5 μl of MgCl2, 5 μl of the oligonucleotide combination at 5 μM, 0.2 μl of Taq Polymerase DNA and 3 μl of genomic DNA at 15 ng μl-1. The PCR was carried out with a thermal cycler (appliedd5’ (ATT CCC ATC TGC ACC ATT) 3’ biosistems®). The profile of the temperature cycles of the PCR was five minutes at 94 °C, followed by 40 cycles consisting of: one minute at 94 °C, one min at the alignment temperatures of 45 to 60 °C and one min and medium at an extension temperature of 72 °C, and finally five more minutes at 72 °C.
The separation of the amplified fragments of DNA was carried out in 6% polyacrylamide gel, by electrophoresis using a vertical chamber UBIBA-K0322SEV at a constant power of 250 V for 1 h and 25 min using 12 μl of the product resulting from the PCR and μl of the 25 bp molecular weight marker (low weight). The resulting gels were stained with ethidium bromide and digitized by a Bio Rad® documentary photo.
With the results of the digitalization and using the CrossChecker software, a binary matrix (1 presence of the band, 0 absence) was built, which was later used to perform the statistical analyzes using the Popgene program version 1.31, calculating for each species and considering all the populations: na or number of alleles observed, ne= number of effective alleles (Kimura and Crow, 1964), h= Nei gene diversity (1973), I= Shannon information index (Lewontin, 1972), number of polymorphic loci, percentage of polymorphic loci, finally with the distance matrix a dendrogram was constructed that presents the diversity found.
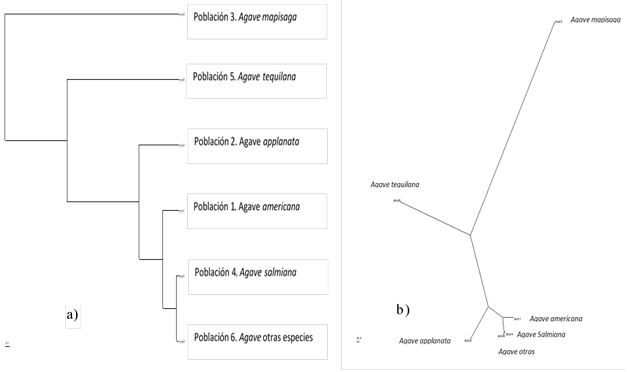
For the genetic diversity analysis subspecies of Agave found in the subprovince of the mountains and plains of northern Guanajuato, were included in the complexes: Agave americana, Agave applanata, Agave mapisaga, Agave salmiana, Agave tequilana and other species where it was included a: Agave desmettiana, Agave weberi and Agave lechuguilla. Taking as a basis the Nei’s unbiased genetic distances calculated for the complexes and using the modified Upgma method (unweighted pair group with Arithmetic Mean) modified from the Neighbor of Phylip procedure version 3.5, a dendrogram and a phylogram of the genetic diversity of agave species were generated in the mountains and plains of northern Guanajuato.
Results and discussion
The taxonomic characterization of agave plants located in the mountains and plains of Guanajuato, allowed the identification of nine species of which four are distributed naturally: Agave salmiana subsp. salmiana, Agave salmiana subsp. crassispina, Agave aplanata and Agave lechuguilla and five have been introduced from different regions of the country: Agave americana subsp. americana, Agave americana spp. protoamericana, Agave mapisaga spp. mapisaga, Agave tequilana and Agave weberi. The results of the taxonomic identification of agave in the mountains and plains of the north of Guanajuato agree with what was found by Herrera, who in his report for the acceptance of San Felipe within the zone of protection by the denomination of origin of mezcal, in addition to the aforementioned reports the species Agave asperrima (possibly referring to A. scabra), Agave tequilana Weber var. azul and Agave angustifolia (IMIPE, 2004).
Although in the present study samples of Agave angustifolia were not collected, it is known that the sale of the cooked quiote of this species is a source of income for some localities in the region, also because of its high sugar content and its wide distribution (Parra et al., 2010) is a ‛wild’ species in several states with mezcal designation of origin, so, strategically for the production of mezcal in Guanajuato, it is advisable to promote the establishment of plantations with this species in the San Felipe and San Luis de la Paz mezcaleros municipalities (Parra, 2018. Com. Per.).
On the other hand, the use of the species of Agave sp. found in the mountains and plains of the north of Guanajuato, according to the settlers, they agree with Shneibar (2008) who mentions that the species Agave americana subsp. americana, Agave salmiana subsp. Crassispina and Agave salmiana subsp. salmiana have been reported with use for mezcal production, possibly due to the fact that the study region is very close to the mezcalera zone of the Potosino highlands where the use of these species is frequent (Aguirre, 2001).
In total, DNA samples were collected from 54 individuals, including those of the species Agave americana var. marginata and Agave desmettiana that were collected in Celaya, Guanajuato to be used as a witness in studies of genetic diversity.
The standardization of modifications of the CTAB protocol modified with STE allowed obtaining DNA concentrations of agave from 100 to 1 000 μg ml-1, with a degree of purity between 1.8 and 2.09 μg ml-1 at a wavelength of A260/A280 nm.
Based on the number of polymorphic bands and their degree of polymorphism, it was determined that the best combinations of ISTR initiators were: F10/B8 and F9/B61 that had already been tested by Torres (2009) in the complex of Agave durangensis, F5/B3 and F6/B3 that were also tested by Montalvo et al. (2010) in seedlings of Pilosocereus sp. and F6/B6II and F5/B6II that were tested in the present study with good results. The best alignment temperatures for these combinations were found at 45, 50 and 60 °C (Table 2).
Table 2 Presence, degree of polymorphism and temperatures of alignment of the combinations of ISTR initiators in agave.
| Combination | TM | Total bands | Polymorphic bands | Polymorphism (%) |
| F6/B6II | 45 | 21 | 19 | 90 |
| F10/B8 | 50 | 18 | 16 | 88 |
| F5/B3 | 50 | 18 | 16 | 88 |
| F9/B61 | 45 | 15 | 13 | 86 |
| F5/B6II | 60 | 23 | 20 | 86 |
| F6/B3 | 50 | 20 | 17 | 85 |
| Total/average | - | 125 | 101 | 87 |
The reproducibility of the ISTR markers in the genus Agave was confirmed, this agrees with what was found by (Eguiarte et al., 1999; Torres et al., 2006; Scheinbar, 2008; Torres et al., 2008; Torres, 2009) as well as reproducibility in the agave genus of the ISTR sequences used in Pilosocereus sp. by Montalvo et al. (2010). Infante et al. (2006) demonstrated in an analysis to discriminate Agavaceae species with ISTR and AFLP markers, that the ISTR markers showed a high degree of polymorphism, as well as the combination F2 and B1A with a high discriminatory power. The combinations showed amplification patterns for the agave species, as well as for the species Agave americana var. marginata used as control DNA (Figure 1).
The results of the diversity analysis (Table 3) indicate that out of a total of 148 amplified loci, from the six combinations of ISTR markers, the polymorphic loci values vary from 42.57% for the Agave tequilana complex to 83.78% for the Agave Salmiana complex. Likewise, the Shannon index for these two complexes varies from 0.2188 for the Agave tequilana complex to 0.3254 for the Agave salmiana complex. The above, indicates that these two complexes are those of greater contrast in the mountains and plains of northern Guanajuato.
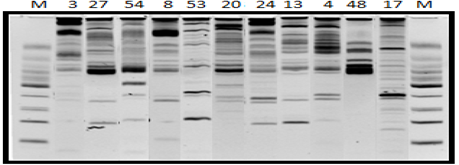
Figure 1 DNA amplification pattern with the combination of ISTR F5/B3 markers for the species: Agave americana spp. Americana (3), Agave americana spp. protoamericana (27), Agave americana var. marginata (54), Agave aplanata (8), Agave desmettiana (53), Agave lechugilla (20), Agave mapisaga spp. mapisaga (24), Agave salmiana spp. crassispina (13), Agave salmiana ssp. salmiana (4), Agave tequilana (48), and Agave weberi (17). The M indicates the Low Weight marker of 25 base pairs.
Table 3 Statistics of gene variation by complex, obtained with 148 loci through the POPGENE program.
| Population Agave | N | na | ne | Gst | I | # Loci polymorphic | (%) Loci polymorphic |
| Americana | 13 | 1.7432 | 1.2888 | 0.191 | 0.3068 | 110 | 74.32 |
| Applanata | 3 | 1. 4324 | 1.2596 | 0.1563 | 0.3251 | 64 | 43.24 |
| Mapisaga | 5 | 1.473 | 1.2525 | 0.1533 | 0.2348 | 70 | 47.3 |
| Salmiana | 20 | 1.8378 | 1.3007 | 0.201 | 0.3254 | 124 | 83.78 |
| Tequilana | 5 | 1.425 | 1.2458 | 0.1449 | 0.2188 | 63 | 42.57 |
| Other | 8 | 1.7703 | 1.3067 | 0.2003 | 0.321 | 114 | 77.03 |
| All | 54 | 1.9662 | 1.3125 | 0.2119 | 0.3474 | 143 | 96.62 |
na= number of alleles observed; ne= number of effective alleles (Kimura and Crow, 1964); h= Nei gene diversity (1973); I= information index of Shannon (Lewontin, 1972).
Similarly, the statistical analysis of the genetic variation by complex (Table 3), indicate that the complex Agave tequilana with a genetic differentiation coefficient (Gst) of Nei (1973) of 0.1449 is the complex with less genetic variation of the total the variability between all the complexes, which would be expected because several of the samples were collected from plantations destined for the industrial production of tequila. On the other hand, the Agave Salmiana complex with 0.201 is the most genetically diverse complex because the subspecies included in this genus are those with the greatest distribution in the sub-province of the mountains and plains of northern Guanajuato, and even those with greater distance among their populations.
In this sense, it is emphasized that the process of domestication of the species, in this case of the Agave, acts as a bottleneck in the loss of genes (polymorphisms) that are necessary to survive in adverse conditions. It is this reason why the Agave tequilana is totally dependent on the hand of man and would have nothing to do in the north of Guanajuato (despite the dream of some agroindustrial), while the conservation and intensive propagation of wild species of Agave as: Agave salmiana ssubsp. crassispina, Agave scabra potosiensis and semi-cultivated species such as Agave salmiana subsp salmiana, Agave mapisaga and Agave americana var. americana (Parra, 2016. Com. Per.).
In response to the growing pressure from the agave resource as a raw material for the production of mezcal in the area with DOM and the existing pressure of “quioteros”, “barbacolleros”, “chiniculieros” and cattle that reduce agave stocks in the region.
On the other hand, the results found using the ISTR markers in agave previous agree with that found by Torres et al. (2012) who using this type of molecular markers in Agave duranguensis reported high levels of polymorphism (24.18 to 61.50%) with 91 amplified loci, with Shannon index of 0.1208 to 0.3435 and genetic diversity (h) from 0.0807 to 0.2337. In addition, the values reported by these authors in their study are similar to those found for the complex other species where they were included: Agave desmettiana, Agave weberi and Agave lechuguilla.
When considering the total of complexes in the genetic diversity analysis (Table 4), a gene differentiation coefficient Gst of 0.1859 is generated, which indicates that the agave complexes of the mountains and plains of northern Guanajuato are poorly differentiated, and that approximately 19% of the variation detected is due to differences between species, the rest 81% represents genetic diversity within the species studied. In addition, the gene flow value (Nm) found between complexes of 2.1892 indicates that the agave complexes of the mountains and plains of northern Guanajuato have a tendency towards homogenization since according to Wright (1931) “a gene flow greater than 1 it leads to the homogenization of the populations”. The above can be influenced, among other things, by anthropogenic issues derived from the types of uses made by the population of the mountains and plains of northern Guanajuato.
Table 4 Analysis of Nei gene diversity in subdivided populations, obtained with 148 loci in the agave species through the POPGENE program.
| Statistical | N | Ht | Hs | Gst | Nm | # Loci polymorphic | (%) Loci polymorphic |
| Means | 54 | 0.2143 | 0.1745 | 0.1859 | 2.1892 | 143 | 96.62 |
| Deviation standard | 0.0213 | 0.014 |
Nm= estimated gene flow from: Gst or Gcs. Eg; Nm= 0.5 (1-Gst)/Gst; (McDermott and McDonald, 1993).
In the original values of Identity and Nei genetic distance generated by the POPGENE program (Table 5) it is observed that the distances between the species studied is very low, possibly it is some variation within the species of Agave salmiana, or, some populations classified as A. salmiana are actually another species such as Agave americana.
Table 5 Original measures of genetic identity and Nei genetic distance, through the POPGENE program.
| Populations Agave | 1 americana | 2 applanata | 3 mapisaga | 4 salmiana | 5 tequilana | 6 others |
| 1 americana | **** | 0.9623 | 0.926 | 0.9859 | 0.9385 | 0.9792 |
| 2 applanata | 0.0384 | **** | 0.8999 | 0.9624 | 0.9172 | 0.9543 |
| 3 mapisaga | 0.0769 | 0.1054 | **** | 0.9304 | 0.8841 | 0.9215 |
| 4 salmiana | 0.0142 | 0.0383 | 0.0721 | **** | 0.9445 | 0.9872 |
| 5 tequilana | 0.0635 | 0.0865 | 0.1232 | 0.0571 | **** | 0.9483 |
| 6 others | 0.021 | 0.0468 | 0.0818 | 0.0129 | 0.0531 | **** |
Genetic identity of Nei are the values above the diagonal and those below is the genetic distance of Nei.
Based on the constructed with Nei’s Neglected genetic distance calculated for the complexes and using the modified UPGMA method of the Phylip Neighbor procedure Version 3.5, a dendrogram and a phylogram of the genetic diversity of agave species were generated in the mountain ranges and plains of the north of Guanajuato, where in both it can be observed that the Agave Salmiana complex is genetically similar to the Agave americana complexes, other agaves and the Agave applanata complex, while with the Agave tequilana and Agave mapisaga complexes it has larger genetic differences, including with the Agave mapisaga complex the genetic differences are very marked (Figure 2).
Conclusions
According to the taxonomic identification using the keys of Gentry 1982, in the mountains and plains of Guanajuato, nine agave species were identified, of which Agave salmiana spp. salmiana, Agave americana spp. americana and Agave salmiana spp. crassispina are the most distributed in this physiographic subprovince.
Six of the 14 combinations of ISTR markers used in the present study showed adequate amplification patterns to study the genetic diversity of the agave species found in the mountains and plains of northern Guanajuato. The agave populations of the mountains and plains of northern Guanajuato are little differentiated. Approximately 19% of the variation detected is due to differences between species, the rest 81% represents genetic diversity within the species studied.
Gratefulness
The authors thank the Council of Science and Technology of the state of Guanajuato (CONCYTEG) for the financing for the execution of the project: identification and genetic diversity of mezcalero agave species in the mountains and plains of northern Guanajuato, which gave rise to the present information.
REFERENCES
Aguirre, R. J. R.; Charcas, S. H. y Flores, F. J. L. 2001. El maguey mezcalero potosino. Universidad Autónoma de San Luis Potosí. Consejo Potosino de Ciencia y Tecnología. San Luis Potosí, SLP, México. 87 p. [ Links ]
Barraza, M. A.; Sánchez, T. F. L.; Robert, M.; Esqueda, M. y Gardea, A. 2006. Variabilidad genética en Agave angustifolia Haw de la Sierra Sonorense, México, determinada con marcadores AFLP. Rev. Fitotec. Mex. 29(1):1-8. [ Links ]
Bellón, M. R.; Barrientos, P. A. F.; Colunga, G. P.; Perales, H.; Reyes, A. J. A.; Rosales, S. R. y Zizumbo V. D. 2009. Diversidad y conservación de recursos genéticos en plantas cultivadas. Capital natural de México, vol. II: estado de conservación y tendencias de cambio. Comisión Nacional para el Conocimiento y Uso de la Biodiversidad (CONABIO). 355-382 pp. [ Links ]
Cuevas, F. X. M. y Flores, B. E. P. 2006. Distancias genéticas entre Agave tequilana Weber var. Azul y especies y variedades afines. Scientia-CUCBA. Guadalajara, Jal. 231-249 pp. [ Links ]
Demey, J. R.; Gámez, E.; Molina, S. and Infante, D. 2004. Comparative study of the discriminating capacity of AFLP and ISTR markers for genetic analysis of Agave fourcroydes. Plant Mol. Biol. Reporter. 22(1):29-35. [ Links ]
IMIPE (Instituto Mexicano de la Propiedad Industrial). 2004. Modificación a la declaración general de protección de la denominación de origen Mezcal. Diario Oficial de la Federación (DOF). [ Links ]
Doyle, J. J. and Doyle, J. L. 1987. A rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochemical Bulletin. 19(1):11-15. [ Links ]
Dreisigacker, S. 2013. Sistemas de marcadores genéticos en el mejoramiento de trigo. In: Reynolds, M.; Pask, A.; Mullan, D. y Chávez, P. (Eds). Fitomejoramiento fisiológico I: enfoques interdisciplinarios para mejorar la adaptación del cultivo. Centro Internacional de Maíz y Trigo (CIMMYT). El Batán, Estado de México. 140-152 pp. [ Links ]
Erazo, P. M. y Cárdenas, R. R. 2013. Ecología impacto de la problemática ambiental actual sobre la salud y el ambiente. Ecoe ediciones. Colombia. 63-85 pp. [ Links ]
Eguiarte, L. E. 1999. Una guía para principiantes a la genética de poblaciones. In: la evolución biológica. Núñez-Farfán J. y Eguiarte, L. E. (Eds.). Facultad de Ciencias, Instituto de Ecología. Universidad Nacional Autónoma de México (UNAM)- Comisión Nacional para el Conocimiento y Uso de la Biodiversidad (CONABIO). 35-50 pp. [ Links ]
Eguiarte, F. L. E. y González, G. A. 2007. De genes y magueyes estudio y conservación de los recursos genéticos del tequila y el mezcal. Ciencias. México, DF. 28-35 pp. [ Links ]
Espinel, S. y Aragonés, A. 1997. Mejora genética de Pinus radiata D. Don. En el país Vasco. Situación actual. Cuadernos de la Sección Forestal. Madrid, España. 5:109-110 pp. [ Links ]
Falcón, L. I. y Valera, A. 2007. Extracción de ácidos nucleicos. In: Eguiarte, L.; Souza, V. y Aguirre, X. (Comps.). Ecología molecular. Instituto Nacional de Ecología (INE)- Secretaría de Medio Ambiente y Recursos Naturales (SEMARNAT) C omisión Nacional para el Conocimiento y Uso de la Biodiversidad (CONABIO). México, DF. 499-516 pp. [ Links ]
García, M. A. J. 2007. Los agaves de México. 2007. Ciencias. México, DF. 14-23 pp. [ Links ]
Garcia-Mendoza, A. 2002. Distribution of the genus Agave (Agavaceae) and its endemic species in México. In: Cactus and Succulent J. 74(4):177-187. [ Links ]
García, M. P. 2014. Diversidad genética de los agaves mezcaleros del municipio de San Felipe, Guanajuato. Tesis ingeniería. División de Agronomía, departamento de botánica. Universidad Autónoma Agraria Antonio Narro (UAAAN). Saltillo, Coahuila. 78 p. [ Links ]
Gámez, V. A. J. 2013. Informe técnico primera etapa del proyecto inventario, potencial productivo y calidad industrial de las especies de agave mezcalero en Guanajuato. INIFAP, Celaya, Guanajuato, México. 45 p. [ Links ]
Gámez, V. A. J. 2014. Informe técnico final del proyecto inventario, potencial productivo y calidad industrial de las especies de Agave mezcalero en Guanajuato. INIFAP, Celaya, Guanajuato, México. 31 p. [ Links ]
Gentry, H. S. A. 1982. Agaves of Continental North America. The University of Arizona Press. Tucson. 670 p. [ Links ]
Godoy, J. A. 2009. La genética, los marcadores moleculares y la conservación de especies. Ecosistemas. 18(1):23-33. [ Links ]
Hernández, S. L.; Pantoja, H. Y. y Martínez, M. 2012. Estudio de caso: plantas útiles y distribución potencial de las forrajeras, medicinales y de uso múltiple. In: Comisión Nacional para el Conocimiento y Uso de la Biodiversidad (CONABIO).La biodiversidad en Guanajuato, estudio de estado Comisión Nacional para el Conocimiento y Uso de la Biodiversidad (CONABIO)-Instituto de Ecología del estado de Guanajuato (IEE). México. 274-289 pp. [ Links ]
Infante, D.; Molina, S.; Demey, J. R. and Gámez, E. 2006. Asexual genetic variability in Agavaceae determined with Inverse sequence tagged-repeats and amplifications fragment length polymorphis analysis. Plant Mol. Biol. Reporter.24(2) Ottawa, Canada. 205-217 pp. [ Links ]
Kimura, M. and Crow, J. F. 1964. The number of alleles that can be maintained in a finite population. Genetics. 49(4):725-738. [ Links ]
Lewontin, R. 1972. The apportionment of human diversity. Evol. Biol. 6(1):381-398. [ Links ]
Mandujano, B. A. 2012. Viabilidad del agave como alternativa para controlar la desertificación de la microcuenca Laguna de Guadalupe, Gto. Tesis Maestría en Ciencias en Gestión Integrada de Cuenca. Universidad Autónoma de Querétaro. Querétaro, Querétaro. 1-32 pp. [ Links ]
Montalvo, F. G.; Ortíz, G. M.; Quiala, M. E.; Keb-Llanes, M.; Rojas, L. E.; Bautista, A. M.; Esquivel, M. A.; Quiroz, M. A.; Rohde, W. y Sánchez, T. L. F. 2010. Primer reporte del empleo de marcadores ISTR en cactaceae (Pilosocereus sp.). Rev. Col. Biot. 12(2):223-229. [ Links ]
Nei, M. 1973. Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences. USA. 70(12):3321-3323. [ Links ]
Parra N. L y Tortolero L A . 2015. Manual del maguey pulquero en Guanajuato. Informe de Investigación de la escuela de desarrollo integral agropecuario de la Universidad Politécnica del estado de Carchi. Tulcán. Ecuador. 25 pp. [ Links ]
Parra, N. L. 2004. Informe final “el centro de propagación de agave del estado de Guanajuato (CEPAEG)”. http://www.intranetfgp.com/siac/2001/111-00/informe%20final/informe%20anual%20cepaeg.doc. [ Links ]
Parra, N. L. 2002. Informe anual de avence “el centro de propagación de agave del estado de Guanajuato (CEPAEG)”. http://www.intranetfgp.com/SIAC/2002/111-00/informe%20final/documentos%20informe%20final/informe%20final.pdf. [ Links ]
Piñero, D. 2008. La diversidad genética como instrumento para la conservación y el aprovechamiento de la biodiversidad: estudios en especies mexicanas. In: Soberón, J.; Halffter, G. y Llorente, J. (Eds). Capital natural de México, vol. I: conocimiento actual de la biodiversidad. Comisión Nacional para el Conocimiento y Uso de la Biodiversidad (CONABIO). México, DF. 437-494 pp. [ Links ]
Rallo, P.; Belaj, A.; De la Rosa, R. y Trujillo, I. 2002. Marcadores moleculares. Córdoba, España. http://www.extremadura21.com/cau-dal/hemeroteca/mayo-junio-2000/almazara/almazara1.htm. [ Links ]
Shneibar, G. E. 2008. Genética de poblaciones silvestres y cultivadas de dos especies mezcaleras: Agave cupreata y Agave potatorum. Tesis de Maestría de la Universidad Nacional Autónoma de México (UNAM). México, D. F. 191 p. [ Links ]
Silvertown, J. W. and Charlesworth, D. 2001. Introduction to plant population biology. Blackwell Science. 125 pp. [ Links ]
Torres, M. M. I.; Almaraz, A. N. and Escoto D. M. 2012. Review article: ISTR, a retrotransposons-based marker to assess plant genome variability with special emphasis in the genera zea and agave. Am. J. Plant Sci. 3(12A):1820-1826. [ Links ]
Torres, M. M. I. 2009. Caracterización molecular del complejo Agave durangensis por medio de marcadores ISTR. Tesis doctoral. Unidad Profesional Interdisciplinaria de Biotecnología, Instituto Politécnico Nacional (IPN). México, D. F. 109 p. [ Links ]
Torres, M. M. I.; Almaraz, A. N.; Velazco, R. A. P.; Hernández, V. V.; Orea, L. G.; Cifuentes, D. A. and Oliver, S. C. 2008. Taxonomic significance of ISTR to discriminate species in Agavaceae. Am. J. Agric. Biol. Sci. 3(4):661-665. [ Links ]
Torres, M. M.; Morales, R. M.; Cruz, L. L. y Villalobos, A. A. 2006. Identificación de polimorfismo entre hijuelos y plantas micropagadas de Agave tequilana y Agave cocui usando ISTR. Scientia Centro Universitario de Ciencias Biológicas Agropecuarias. Universidad de Guadalajara. México. 8(2):203-206. [ Links ]
Wright, S. 1931. Evolution in Mendelian populations. Genetics. 16(2):97-159. [ Links ]
Received: January 00, 2018; Accepted: March 00, 2018











 texto en
texto en