Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO
Share
Revista mexicana de ciencias agrícolas
Print version ISSN 2007-0934
Rev. Mex. Cienc. Agríc vol.9 n.3 Texcoco Apr./May. 2018
https://doi.org/10.29312/remexca.v9i3.267
Articles
Presence of Candidatus Liberibacter solanacearum in Bactericera cockerelli Sulc associated with diseases in tomato, chili and potato
1Universidad Autónoma de Sinaloa-Doctorado en Biotecnología Programa Regional del Noroeste de la Facultad de Ciencias Químico Biológicas. Av. Las Américas y Josefa Ortiz, Culiacán, Sinaloa, México. CP. 80030.
2Universidad Autónoma de Sinaloa-Facultad de Medicina Veterinaria y Zootecnia. San Ángel núm. 3886, Fraccionamiento San Benito, Culiacán, Sinaloa, México. CP. 80260.
3Campo Experimental Valle de Culiacán-INIFAP. Carretera Culiacán-Eldorado km 17.5, Culiacán. Sinaloa, México. CP 80000.
The “permanent tomato”, “spotted tuber” or “zebra chip” in potato and “chlorotic buds” of chili, are three diseases described in Mexico with coincidental signs of flower abortion, obscuration of vascular tissue at the base of the stem and root of the plants. The association between these diseases and the emerging bacterium Candidatus Liberibacter solanacearum (CLs) as well as the psyllid Bactericera cockerelli as its vector has been mentioned. These diseases, which were initially located in three states of Mexico, have spread to the main Solanaceae producing regions, both in field conditions and in the greenhouse. The objective of the study was to know the presence of CLs associated with diseases that affect the tomato, para and chili crops in Mexico. The bacterium was identified by PCR of the 16S rDNA gene, cloning and sequencing. The alignment of nucleotide sequences was performed with the Clustal W method and the phylogenetic tree was built with the Neighbor-Joining algorithm from distances calculated with the Tajima-Nei method and a Felsenstein index of 1 000 replicas, using the software MEGA version 5.05. A total of 167 samples were analyzed, of which 86 were positive, from 14 states of Mexico. Five nucleotide sequences from Guanajuato, San Luis Potosi and Sinaloa corresponding to tomato, potato and chili, adults and eggs of B. cockerelli were obtained. The analysis of the sequences showed an identity of 99.4% when compared between these and up to 99.8% with GenBank accessions described for CLs in the USA, New Zealand and Canada.
Keywords: chlorotic buds; tomato permanent; zebra chip
El “permanente del tomate”, “manchado del tubérculo” o “zebra chip” en papa y “brotes cloróticos” del chile, son tres enfermedades descritas en México con signos coincidentes de aborto de flor, oscurecimiento de tejido vascular en la base del tallo y raíz de las plantas. Se ha mencionado la asociación entre estas enfermedades y la bacteria emergente Candidatus Liberibacter solanacearum (CLs) así como al psílido Bactericera cockerelli como su vector. Estas enfermedades, que en inicio se localizaban en tres estados de México se han diseminado a las principales regiones productoras de solanáceas, tanto en condiciones de campo como en invernadero. El objetivo del estudio fue conocer la presencia de CLs asociado a enfermedades que afectan los cultivos de tomate, para y chile en México. La bacteria se identificó por PCR del gen 16S de ADNr, clonación y secuenciación. La alineación de secuencias nucleotídicas se realizó con el método Clustal W y el árbol filogenético se construyó con el algoritmo de Neighbor-Joining a partir de distancias calculadas con el método de Tajima-Nei y un índice de Felsenstein de 1 000 réplicas, utilizando el software MEGA versión 5.05. En total se analizaron 167 muestras, de las cuales 86 resultaron positivas, provenientes de 14 estados de México. Se obtuvieron cinco secuencias nucleotídicas de Guanajuato, San Luis Potosí y Sinaloa correspondientes al tomate, papa y chile, adultos y huevecillos de B. cockerelli. El análisis de las secuencias mostró una identidad de 99.4% al comparase entre estas y hasta 99.8% con accesiones del GenBank descritas para CLs en EUA, Nueva Zelanda y Canadá.
Palabras clave: brotes cloróticos; permanente del tomate; zebra chip
Introduction
In Mexico, there are three diseases that have caused major changes in the crop program in the country’s geography. The “permanent tomato disease” (PT) was first described in the Bajio region of Guanajuato causing damage to tomato production (Solanum lycopersicum L.) estimated at 45%, later, in the southern states of Coahuila and Nuevo León mentioned a new disease in the potato crop (Solanum tuberosum L.) named “Zebra chip” (ZC) causing 90% damage (Munyaneza et al., 2009b) and in chili a new disease described as “chlorotic buds” (BC) (Munyaneza et al., 2009c).
The symptoms of PT are characterized by a curling of the lower leaves in the form of “taco”, these leaves are brittle, the apical leaflets are chlorotic with purple margins, the flowers are aborted; nevertheless, it is possible to present an over budding of buds that form thin stems that bloom and reach the production of small fruits. In general, the plants are squashed with yellow leaves and the vascular tissue in the root has a dark brown color (Garzón-Tiznado et al., 2009). In the case of potatoes, the symptoms of the ZC are characterized by purple to chlorotic buds, bulky axillary stems and radial obscuration of the tuber, which loses frying quality and its cooking is nonuniform, these potatoes have a slightly sweet taste, with which they lose their commercial quality.
Finally, in bell-type chili the symptoms of BC are shortening of internodes and small plants (Munyaneza et al., 2009c). Results of initial investigations suggested that these diseases were caused by the same pathogen, since when grafting tomato plants diseased with PT in healthy potato plants they showed “purple tip” symptoms. Later, it was mentioned that the symptoms in tomato (Garzón-Tiznado et al., 2009) and potatoes were reproduced by the psyllid B. cockerelli.
Initially, PT and ZC diseases were associated with phytoplasmas; however, when analyzing the DNA sequence of the 16S-23S region amplified with the pairs of direct primers P1/Tint and P1/P7 and nested R16mF2/r16r2 described for phytoplasmas (Gundersen and Lee, 1996), they presented an identity of 80 to 83% when compared to sequences described for phytoplasmas. In contrast, these sequences revealed a greater association with a non-culturable bacterium transmitted by B. cockerelli (Garzon-Tiznado et al., 2009) that was later described with 100% identity with the bacterium Candidatus Liberibacter solanacearum in tomato (Munyaneza et al., 2009a) and in chili cultivation (Munyaneza et al., 2009c) in the Valley of the Cruz of Elota, Sinaloa.
However, the PT was detected in the early 80’s and later the spotting of the potato tuber (Almeyda et al., 2008), characteristic signs of these diseases have been observed in tomato, chili and potato plantations, in the Mexico's main producing regions, so it was considered important to obtain information on the presence of these diseases in Mexico and their relationship with the bacterium Candidatus Liberibacter solanacearum.
Materials and methods
Inoculum of Candidatus Liberibacter solanacerum. Under conditions of commercial greenhouse and open field, foliar tissue of tomato plants that had symptoms of “PT”, as well as of potato with symptoms of “ZC” and of chili with “BC” was collected, eggs were also collected, nymphs and B. cockerelli adults who colonized tomato plants with PT. The plant samples were placed in plastic bags and stored at -20 °C. The adults of the psyllid were collected with entomological sniffer and the eggs and nymphs were removed from the leaflets with “camel hair” brush and placed in 1.5 mL tubes in 75% ethanol, washed with sterile distilled water and stored at -20 °C.
Extraction of DNA. The extraction of nucleic acids from plants and insects, eggs and nymphs of B. cockerelli was carried out with the methodology described by Doyle and Doyle (1990) modified. The 500 mg of foliar tissue was macerated in mortar with 2 mL of extraction solution (100 mM Tris-HCl pH7, 2% CTAB, 20 mM EDTA, 1.4 M NaCl and 0.2% β-mercaptoethanol). In the DNA extractions of the B. cockerelli stages, 100 mg of sample was macerated with 200 μL of extraction solution. The extracts were incubated at 65 °C for 30 min. After ammonium acetate was added to a final concentration of 0.5 M and incubated on ice for 10 min, a volume of phenol: chloroform: isoamyl alcohol solution (25: 24: 1 v/v/v) was added and stirred in vortex, centrifuged at 13.4 x g for 10 min, the supernatant was recovered and 0.6 volumes of isopropanol were added, the DNA was allowed to precipitate for 1 h at -20 °C. Subsequently, it was centrifuged at 13.4 x g for 10 min. The supernatant was removed and the precipitate was washed in 500 μL of 70% ethanol, allowed to dry and suspended in 50 μL (plant samples) and 20 μL (B. cockerelli stage samples) of sterile distilled water. The extracted DNA was stored at 4 °C.
Detection of CLs by PCR. For the detection of CLs, the pair of OA2/OI2c primers was used, which amplify a 1 168 bp fragment of the 16S rDNA gene (Munyaneza et al., 2009), in a 25 μL reaction volume containing 3 mM of MgCl2, 0.2 mM of DNTPs, 0.25 μM of each oligonucleotide, 1 U of Taq DNA polymerase (Invitrogen) and 50 ng of DNA. The reaction was carried out in a C1000TM Thermal Cycler BIO-RAD thermal cycler, with pre-denaturation of 2 min at 94 ºC followed by 35 cycles at three temperatures: denaturation at 94 ºC for 30 s, alignment at 60 ºC for 30 s and extension at 72 ºC for 1 min; with a final period of 10 min at 72 ºC. The amplicons were analyzed on 1.2% agarose gels stained with gel network (BIOTIUM) and observed in UV light.
DNA cloning and sequencing. The PCR-amplified products were purified from agarose gels using commercial mini-columns (PureLinkTM) and cloned into the pCR 4-TOPO vector following the supplier's instructions (Invitrogen). Plasmid DNA extraction was performed with the method described by Birnboim and Doly (1979). The transformed bacteria were confirmed by restriction bioassays with the EcoRI enzyme following the supplier’s instructions (Invitrogen). The sequencing was performed in LANGEBIO, CINVESTAV-IPN- Irapuato in a 3730 XL DNA sequencer (applied Biosystems, Foster City, CA, USA). The samples were prepared using the BigDye Terminator v3.1 sequencing kit (Applied Biosystems). The DNA identity was obtained by comparing the sequences obtained in this study with those reported in the NBCI database (National Center for Biotechnology Information) using the BLAST algorithm (Basic Local Alignment Search Tool), (Altschul et al., 1990).
Phylogenetic analysis. The alignments of the nucleotide sequences of CLs were made with the Clustal W method and the phylogenetic tree was constructed with the Neighbor-Joining method (Saitou and Nei, 1987) from distances calculated with the Tajima-Nei model (Tajima and Nei, 1984) and a bootstrap of 1 000 replicas (Felsenstein, 1985). Everything analyzes were performed with the MEGA program version 5.05 (Tamura et al., 2011).
Results and discussion
Detection of CLs by PCR. In 167 samples collected in 14 georeferenced states of the Mexican Republic, 86 samples were amplified with a predicted size for CLs of 1 168 bp. In tomato samples the detection of CLs was 62%, in potatoes 40.3%, 33% in the chili culture, as well as 100% in adults, eggs and nymphs of B. cockerelli collected in diseased tomato plants (Table 1). In all three cultures, the common symptom was characterized by flower abortion and brownish darkening at the base of the stem and root of the plant coinciding with the previously described symptoms (Garzón-Tiznado et al., 2009; Munyaneza et al., 2009c).
Table 1 Location of samples of tomato (Solanum lycopersicum L.), potato (Solanum tuberosum L.) and chili (Capsicum annuum L.) analyzed by PCR, for the detection of Candidatus Liberibacter solanacearum in Mexico.
| State | Coordinates | CLs positives | ||||
| S. lycopersicum | S. tuberosum | C. annuum | B. cockerelli * | B. cockerelli ** | ||
| Aguascalientes | 22º05’13.00” N 102º42’25.30” O | 3/5 | 1/5 | 1/1 | 1/1 | - |
| Baja California | 30°25’54.81” N 115°52’55.81” O | 1/10 | - | - | - | - |
| Chihuahua | 28º19’1.85” N 106º56’69.00” O | - | 1/4 | - | - | - |
| Coahuila | 25º19’20.76” N 100º39’56.44” O | - | 7/10 | - | - | - |
| Estado de México | 19º05’42.46” N 99º34’23.15” O | - | 4/8 | - | - | - |
| Guanajuato | 20°39’48.17” N 100°45’43.62” O | 8/8 | - | 1/5 | 4/4 | 1/1 |
| Guanajuato | 21°3’11.66” N 101°42’1.28” O | - | 4/8 | - | - | - |
| Jalisco | 19º52’35.81” N 103º44’32.83” O | - | 1/5 | - | - | - |
| Jalisco | 19º45’12.16” N 103º45’36.08” O | 5/5 | - | - | - | - |
| Michoacán | 19º48’44.88” N 102º22’25.09” O | - | 1/5 | - | - | - |
| Michoacán | 20º19’23.52” N 102º18’19.23” O | 1/2 | - | - | - | - |
| Nayarit | 21º45’59.54” N 105º13’58.27” O | 1/5 | - | - | - | - |
| Nayarit | 21º42’21.61” N 105º16’41.75” O | - | - | 0/2 | - | - |
| Nuevo León | 24º34’24.14” N 100º16’34.27” O | - | 4/7 | - | - | - |
| San Luis Potosí | 22°39’56.30” N 100°53’38.50” O | 5/5 | - | 1/1 | - | - |
| Sinaloa | 23°57’11.73” N 106°52’18.99” O | 6/6 | - | 5/5 | 3/3 | 3/3 |
| Sinaloa | 23º54’57.68” N 106º53’3.91” O | - | 3/5 | - | - | - |
| Sonora | 27º21’40.59” N 110º14’30.93” O | - | 2/2 | - | - | - |
| Sonora | 26º51’51.40” N 109º32’0.10” O | 1/1 | - | - | - | - |
| Zacatecas | 23º1’28.37” N 100º39’56.44” O | 5/11 | 1/13 | 0/10 | 1/1 | - |
| Total | 36/58 (62%) | 29/72 (40.27%) | 8/24 (33.33%) | 9/9 (100%) | 4/4 (100%) | |
*= adults; **= eggs.
In tomato cultivation, the highest frequency of the pathogen was detected; the symptoms of brittle leaves, chlorotic apical leaflets with purple margins, in addition to the darkening of the roots and abortion of flowers, allowed during the development of the crop the selection of plants where the presence of the pathogen was confirmed (Figure 1).
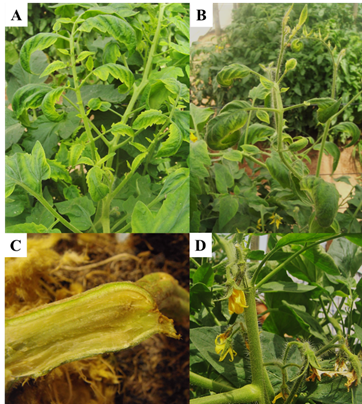
Figure 1 Symptoms associated with CLs in tomato plants with PT. A) chlorotic apical leaflets and epinastia; B) on sprouting of buds and chlorotic leaflets with purple margins; C) brown color in vascular tissue of the root; D) flower abortion.
In potato cultivation with a lower frequency, the purple tip syndrome has been described as complex in nature and can also be induced by pathogens other than CLs (Garzón-Tiznado et al., 2008). However, the presence of the pathogen is also associated with the symptom of stem necrosis and zebra chip, which occurs from the intermediate stage of crop development, which made it difficult to detect it in early samples. In the culture of the chili in samplings realized in the states of Nayarit and Zacatecas, the presence of CLs was not detected, in coincidence with the absence of symptoms of chlorotic buds in the plants sampled at random in genotypes of type ‛wide’ or ‛serrano’; however, in the ‛bell’ or ‛habanero’ types of samples collected in Guanajuato, Sinaloa and Aguascalientes, the previously described symptoms were observed and the results for the detection of CLs were positive.
The above, suggests that the symptoms can be a first-hand tool for the support of technicians in the field, although the confirmation and certainty of the identity of the bacteria can only be done in the laboratory. In addition, CLs were identified in samples of adults, eggs and nymphs of B. cockerelli that hosted tomato plants with symptoms of PT and that were positive for CLs (Figure 2A). The presence of CLs was detected in potato and tomato producing states in Mexico (Table 1). In Sinaloa, Sonora, Nayarit and Jalisco with prevalence of tropical climate and plantings in winter, the damage caused by this pathogen has not yet been as critical as in the other states, where the temperate climate and sowing in summer predominates.
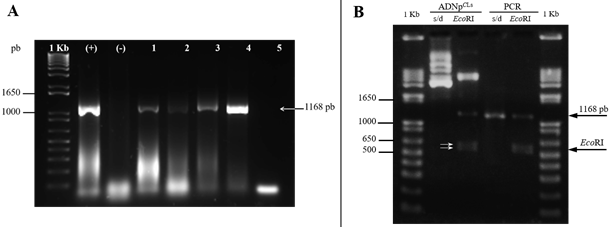
Figure 2 Amplicons of the 16S gene of CLs and restriction of the recombinant plasmid with EcoRI. A) detection of CLs by PCR. In lanes 1 and 2 positive and negative controls respectively. Lanes 3, 4 and 5 correspond to eggs, nymphs and adults of B. cockerelli. Lane 6, tomato plant with PT and lane 7, negative control of the PCR reaction. B), restriction pattern of plasmid p CLs . From left to right, plasmid DNA from B. cockerelli clone and plasmid digested with EcoRI. Then, amplicon, amplicon digested with EcoRI and marker of molecular weight of 1 Kb. Electrophoresis in 1.5% agarose gel.
The tomato permanent disease is a disease described in the early 80s in the state of Guanajuato, from there has been observed its spread to other regions of the country. Currently, the morphological characteristics of this disease have been registered in the main potato producing regions in Mexico and those where it is combined simultaneously with the tomato crop. Similar morphological characteristics have also been described in countries such as the United States of America (French-Monar et al., 2010) and New Zealand (Liefting et al., 2009).
DNA cloning and sequencing. Amplicons obtained from DNA extracted from adults and eggs of B. cockerelli, as well as from tomato, potato and chili cloned in the vector pCR 4-TOPO, were confirmed by partial restriction with the enzyme EcoRI, which allowed the release of two fragments of DNA that added corresponds approximately to the size of the amplicon, which indicated the presence of an intermediate EcoRI site (Figure 2B, data shown for psyllids of B. cockerelli), which was confirmed by sequencing. The sequences obtained in this study were registered in GenBank with the numbers KF776420 and KF776421 for CLs of adults and eggs of B. cockerelli respectively, KF776422 for CLs in tomato with symptoms of PT, KF776423 for CLs in potatoes with symptoms of ZC and KF776424 and for CLs in chili with symptoms of BC.
Analysis of the 16S gene sequences of CLs. Both in the sequences reported in this study, and those recorded in the GenBank used in the analyzes they showed an EcoRI site from position 616 to 621 which was observed in the restriction pattern of the recombinant plasmid with EcoRI (Figure 2B). To locate the position of the mutations and the EcoRI sites, the sequence of the complete 16S rRNA gene of 1507 bp of the Candidatus Liberibacter solanacearum genome (NC_014774) was used as reference.
The nucleotide sequences of tomato tissue amplicons with PT, potato with ZC, chili with BC and, eggs and adults of B. cockerelli collected in the tissue of diseased tomato, potato and chili plants described in this study presented an identity of 99.4% when comparing each other; that is, a variation of 0.6% was presented. The comparison between sequences previously described for CLs from New Zealand, Texas and Kansas from USA and Sinaloa, Saltillo and Chihuahua from Mexico presented 99.8% identity, 98.5% with Ca. Liberibacter africanus, 98.4% with Ca. Liberibacter asiaticus, 97.7 with Ca. Liberibacter americanus, 97.2% with Ca. Liberibacter europaeus, data similar to those previously described by Liefting et al. (2009) and 93.1% with Ca. Phytoplasma, which showed an even greater difference (Table 2). With the above, the tendency of a greater distance of the PT, ZC, BC with Ca. Phytoplasma is evident.
Although in the beginning PT was associated with phytoplasmas, the results of the molecular analysis confirmed a greater association with CLs similar to that described by Munyaneza et al. (2009). The adult sequence of Bactericera cockerelli showed greater nucleotide variation, among them three mutations were identified: a deletion at position 475 (-/A), a transversion at position 559 (C/G) and a transition at position 822 (A/G). These variations suggest the presence of haplotypes of the bacteria, as has been reported in studies with Bactericera cockerelli, potato and tomato samples from the United States of America, New Zealand and Mexico (Warrick et al., 2011).
To date, five haplotypes of CLs have been reported, haplotypes A and B associated with diseases transmitted by Bactericera cockerelli in Solanaceous species, and haplotypes C, D and E, associated with diseases in apiaceas (carrots and celery), transmitted by Trioza aplicalis and Bactericera trogonica, as well as the low probability that Bactericera cockerelli transmit CLs to carrots under field conditions (Munyaneza et al., 2016).
Table 2 Comparison of percentage identity between sequences of tomato (S. lycopersicon L.), potato (S. tuberosum L.), chili (Capsicum annuum L.) and Bactericera cockerelli Sulc, reported in this study with sequences described in GenBank of species of Ca. Liberibacter and Ca. Phytoplasma
| GenBank | Host | % Identity | Pathogen | Host | GenBank(Others studies) |
| KF776422 | S. lycopersicum | 99.8 | Ca. Liberibacter | S. lycopersicum | FJ829812, FJ829811, |
| KF776423 | S. tuberosum | solanacearum | S. tuberosum | FJ498806, NC-014774, | |
| KF776424 | C. annuum | C. annuum | GQ468843, JF811596, | ||
| KF776421 | B. cockerelli | JF811597, FJ939136, | |||
| (Huevecillos | FJ939137, FJ957897, | ||||
| KF776420 | B. cockerelli | EU834130, EU918197, | |||
| FJ957896 | |||||
| 98.5 | Ca. Liberibacter | Trioza erytreae | L22533 | ||
| africanus | Citrus sp | EU921620 | |||
| 98.4 | Ca. Liberibacter | Diaphorina citri | GQ502291, JX430434 | ||
| asiaticus | Citrus sp | AB480089, DQ302750, | |||
| DQ71900 | |||||
| 97.7 | Ca. Liberibacter | Diaphorina citri | AY742824 | ||
| americanus | |||||
| 97.2 | Ca. Liberibacter | Cacopsylla pyri | FN678792, JX244258 | ||
| europaeus | |||||
| 93.1 | Ca. Phytoplasma | S. lycopersicum | JX311953, L33760, | ||
| AY180955 |
The high identity between the sequences corresponding to eggs and adults of B. cockerelli, of diseased tissue of tomato, potato and chili in this study, allows us to confirm B. cockerelli as a transmitter of CLs, previously described as “non-culturable bacteria” (Garzón-Tiznado et al., 2009), associated with diseases PT, ZC and BC, which demonstrates the epidemiological importance of the relationship between these organisms, and CLs placing this pathogen as responsible for large economic losses recorded in crops such as potatoes and tomato in Mexico (Garzón-Tiznado et al., 1992; Garzón-Tiznado et al., 2009; Munyaneza et al., 2009). In addition, the identification of CLs in eggs of B. cockerelli supports the hypothesis of vertical or transovarial transmission of this pathogen.
Phylogenetic analysis. In Figure 3, the phylogeny corresponding to the sequences of tomato, potato, chili, adults and eggs of B. cockerelli is observed, the sequences analyzed in this study are part of the sequence group of Ca. Liberibacter, species solanacearum with 99% of reproducibility based on the Felsestein index of 1000 replicas and separated from other species of Ca. Liberibacter and Ca. Phytoplasma.
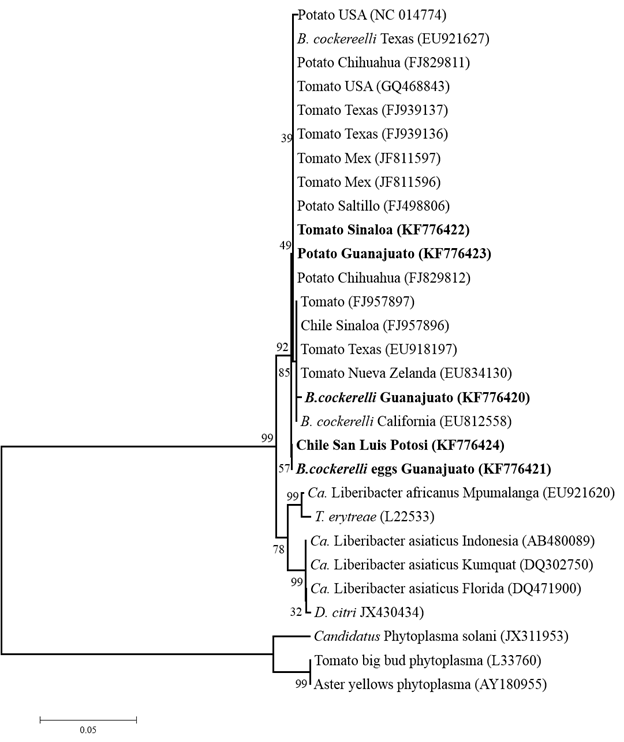
Figure 3 Phylogram based on the sequence comparison of the 16S gene of Candidatus Liberibacter and Ca. Phytoplasma species. The phylogenetic tree was constructed with the Neighbor-Joining algorithm from distances calculated using the Tajima-Nei method, analyzing a total of 992 positions. In bold, the sequences of CLs obtained in this study are identified.
Conclusions
The presence of Candidatus Liberibacter solanacearum was detected in the 14 sampled states of the Mexican Republic in samples of DNA extracted from tomato, chili and potato tissue, eggs, nymphs and adults of Bactericera cockerelli.
The presence of the bacteria was detected more frequently in tomato plants (62%), followed by potatoes (40.27%) and in a lower percentage in chili plants (33.33%).
Phylogenetic analyzes indicate the relationship between diseases of “tomato permanent” (PT), “zebra chip” (ZC) in potato, and “chlorotic outbreaks” (BC) in Chili with the non-culturable bacterium Candidatus Liberibacter solanacearum.
The presence of CLs in samples of eggs of B. cockerelli suggests that the transmission of the bacteria is transovarial.
Literatura citada
Almeyda, I. H.; Sánchez, J. A and Garzón-Tiznado J. A. 2008. Vectors causal of potato purple top in Coahuila and Nuevo León, México. Agric. Téc. Méx. 34(2):141-150. [ Links ]
Altschul, S. F.; Gish, W.; Miller, W.; Myers, E. W. and Lipman, D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [ Links ]
Birnboim, H. C. and Doly, J. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7(6):1513-1523. [ Links ]
Doyle, J. J. and Doyle, J. L. 1990. Isolation of plant DNA from fresh tissue. Focus. 12:13-15. [ Links ]
Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 39:783-791. [ Links ]
French-Monar, R. D.; Patton III, A. F. and Douglas J. M. 2010. First report of “Candidatus Liberibacter solanacearum” on field tomatoes in the United States. Plant Pathol. 94(4):481. [ Links ]
Garzón, J. A.; Becerra, F. A.; Marín, A.; Mejía, A. C. y Byerly, K. F. 1992. Manejo integrado de la enfermedad “permanente del tomate” (Lycopersiconly copersicum Karst ex FawllMill.), en el Bajío. In: afidos como vectores de virus en México. Urías, C.; Rodríguez, R. y Alejandre, T. (Ed.). Colegio de Postgraduados, Centro de Fitopatología. México. 116-129 pp. [ Links ]
Garzón, J. A.; Bujanos, M. R.; Velarde, F. S.; Parga, V.; Avilés, M. C.; Almeida, I. H.; Sánchez, A.; Martínez, J. L. y Garzón, C. J. A. 2008. Bactericera (Paratrioza) cockerelli Sulc, vector de fitoplasmas en México. Ediciones Parnaso, España. In: detección, diagnóstico y manejo de la enfermedad punta morada de la papa. 135 p. [ Links ]
Garzón, J. A.; Cárdenas, O. G.; Bujanos, R.; Marín, A.; Becerra, A.; Velarde, S.; Reyes, C.; González, M. y Martínez, J. L. 2009. Asociación de Hemiptera: Triozidae con la enfermedad “permanente del tomate” en México. Agric. Téc. Méx . 35(1):61-72. [ Links ]
Gundersen, D. E. and Lee, I. M. 1996. Ultrasensitive detection of phytoplasmas by nested-PCR assays using two universal primer pairs. Phytopathol. Mediterranean. 35(3):144-151. [ Links ]
Liefting, L. W.; Sutherland, P. W.; Ward, L. I.; Paice, K. L.; Weir, B. S. and Clover, R. G. 2009. A new “Candidatus Liberibacter” species associated whit diseases of solanaceous crops. Plant Dis. 93(3):208-214. [ Links ]
Liefting, L. W.; Weir, B. S.; Pennycook, S. R. and Clover, R. G. 2009. “Candidatus Liberibacter solanacearum”, a Liberibacter associated with plants in the family Solanaceae. Inter. J. Systematic Evol. Microbiol. 59:2274-2276. [ Links ]
Liefting, L. W.; Pérez, Z. C.; Clover, R. G. and Anderson, A. D. 2008. A new “Candidatus Liberibacter” species in Solanum tuberosum in New Zealand. Plant Dis. 92:1474-1480. [ Links ]
Munyaneza, J. E.; Mustafa, T.; Fisher, T. W.; Sengoda, V. G. and Horton, D. R. 2016. Assessing the likelihood of transmission of Candidatus Liberibacter solanacearum to carrot by potato psyllid, Bactericera cockerelli (Hemiptera: Triozidae). PLoS One. Doi: 10.1371/journal.pone.0161016. [ Links ]
Munyaneza, J. E.; Sengoda, V.G.; Crosslin, J. M.; Garzón, J. A. and Cárdenas, O. G. 2009c. First report of Candidatus Liberibacter solanacearum in pepper plants in México. Plant Dis. Doi:10.1094/PDIS-93-10-1076B. [ Links ]
Munyaneza, J. E.; Sengoda, V. G.; Crosslin, J. M.; De la Rosa, G. and Sánchez, A. 2009b. First report of “Candidatus Liberibacter psyllaurous” in potato tubers with zebra chip disease in Mexico. Plant Dis . Doi:10.1094/PDIS-93-5-0552A. [ Links ]
Munyaneza, J. E.; Sengoda, V. G.; Crosslin, J. M.; Garzón, J. A. and Cárdenas, O. G. 2009a. First report of Candidatus Liberibacter solanacearum in tomato plants in México. Plant Dis . Doi:10.1094/PDIS-93-10-1076A. [ Links ]
Saitou, N. and Nei, M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [ Links ]
Tajima, F. and Nei, M. 1984. Estimation of evolutionary distance between nucleotide sequences. Mol. Biol. Evol. 1:269-285. [ Links ]
Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M. and Kumar, S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731-2739. [ Links ]
Warrick, W. R.; Fisher, T. W. and Munyaneza, J. E. 2011. Haplotypes of “Candidatus Liberibacter solanacearum” suggest long-standing separation. Eur. J. Plant Pathol. 130:5-12. [ Links ]
Received: March 00, 2018; Accepted: May 00, 2018











 text in
text in 


