Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Similares em
SciELO
Similares em
SciELO
Compartilhar
Revista mexicana de ciencias agrícolas
versão impressa ISSN 2007-0934
Rev. Mex. Cienc. Agríc vol.9 no.1 Texcoco Jan./Fev. 2018
https://doi.org/10.29312/remexca.v9i1.844
Articles
Identification of Mexican lemon hybrids using molecular markers SSR
1Colegio de Postgraduados-Campus Montecillo. Carretera México-Texcoco km 36.5. CP. 56230. Texcoco, Estado de México. México. Tel. 01(595) 9520200. (alexge@colpos.mx; sercruz@colpos.mx).
2Campo Experimental Tecomán-INIFAP. Carretera Colima-Manzanillo km 35. Tecomán, Colima, México. CP. 28930. Tel. 01 (800) 0882222, ext. 84332. (robles.manuel@inifap.gob.mx).
The development of tolerant varieties can help to confront and reduce the impacts of diseases that threaten the cultivation of Mexican lemon [Citrus aurantifolia (Christm) Swingle]. Hybrid interspecific and intergeneric plants have been generated in search of genotypes with resistance or greater tolerance to VTC, HLB and anthracnose. Because most citrus fruits are apomictic and polyembryonic the identification of hybrid plants is required by techniques such as molecular genetic markers. The objective of the study was to identify Mexican lemon hybrids using molecular markers called microsatellites (SSR). A population of 203 individuals of crosses carried out from 2009 to 2012 was used in the INIFAP-Tecoman Experimental Field. The genomic DNA was made according to the CTAB 2% method. The PCR reaction was done with a volume of 20μl containing 10 μl of iTaq™ Universal Probes supermix and 2.5 μl of each Forward and Reverse primer. The primers used were TAA41, TAA45, cAGG9 and TAA52. The amplified products were analyzed in 8% polyacrylamide gel and stained with 0.2% silver nitrate. The identification of hybrid plants was carried out by comparing the bands produced by the hybrids with those of the parents. The four initiators allowed the identification of hybrid plants. The highest number of identified plants was obtained with primers TAA45 and cAGG9 with 145 and 130 respectively. The identification of the hybrids of the population under study through the analysis of molecular markers, will allow genetic improvement programs to be efficient, which implies obtaining a large number of seedlings derived from the crosses made.
Keywords: Citrus aurantifolia; anthracnose; HLB; microsatellites; VTC
El desarrollo de variedades tolerantes puede ayudar a enfrentar y reducir los impactos de enfermedades que amenazan al cultivo de limón mexicano [Citrus aurantifolia (Christm) Swingle]. Se han generado plantas hibridas interespecíficas e intergenéricas, en busca de genotipos con resistencia o mayor tolerancia a VTC, HLB y antracnosis. Debido a que la mayoría de los cítricos son apomícticos y poliembriónicos se requiere la identificación de plantas híbridas por medio de técnicas tales como los marcadores genéticos moleculares. El objetivo del estudio fue identificar híbridos de limón mexicano mediante marcadores moleculares denominados microsatélites (SSR). Se utilizó una población de 203 individuos de cruzas realizadas de 2009 a 2012, en el Campo Experimental Tecomán-INIFAP. El ADN genómico se realizó de acuerdo con el método CTAB 2%. La reacción de PCR se hizo con un volumen de 20µl conteniendo 10 µl de iTaq™ Universal Probes supermix y 2.5 µl de cada iniciador Forward y Reverse. Los iniciadores utilizados fueron TAA41, TAA45, cAGG9 y TAA52. Los productos amplificados, se analizaron en gel de poliacrilamida al 8% y se tiñeron con nitrato de plata al 0.2%. La identificación de plantas hibridas se realizó mediante la comparación de bandas producidas por los híbridos con los progenitores. Los cuatro iniciadores permitieron la identificación de plantas híbridas. El mayor número de plantas identificadas se obtuvieron con los iniciadores TAA45 y cAGG9 con 145 y 130 respectivamente. La identificación de híbridos de la población en estudio a través del análisis de marcadores moleculares, permitirá que los programas de mejoramiento genético sean eficientes, que implica la obtención de un gran número de plántulas derivadas de las cruzas realizadas.
Palabras clave: Citrus aurantifolia; antracnosis; HLB; microsatélites; VTC
Introduction
In recent years, the cultivation of Mexican lemons has been seriously threatened by the arrival of diseases that threaten their permanence as the most important agricultural activity for the state of Colima and of great relevance for the states of Michoacán, Guerrero and Oaxaca. The fungus-type diseases such as anthracnose (Colletotrichum acutatum) stand out, occur during the rainy season and affect the fruit's yield and quality in the autumn-winter months. Citrus tristeza virus (VTC), viral disease of major economic importance affecting the cultivation of citrus fruits worldwide and huanglongbing (HLB) currently considered the most devastating being more dangerous than the VTC.
The development of tolerant varieties can help to confront and reduce the impacts of these diseases. The Mexican lemon breeding program of the INIFAP-Tecoman has generated interspecific and intergeneric hybrid plants, in the search for genotypes with resistance or greater tolerance to these diseases and in this way to preserve the Mexican lemon crop in the country.
The programs of genetic improvement of citrus fruits in several parts of the world, are focused mainly to the generation of varieties of fruit without seed, production of genotypes for use as rootstocks that integrate besides good compatibility with the varieties, tolerance to pathogens and adaptation to soils with salinity and calcium carbonates (Tusa et al., 1996; Grosser et al., 2000; Ollitrault et al., 2000; Cristofani et al., 2002; Navarro et al., 2002; Grosser and Gmitter, 2005; Shi-Ping et al., 2009).
Some reports indicate that it is possible to obtain triploid segregates (3x) by crossing diploid citrus genotypes (2x) with tetraploid genotypes (4x) (Cameron and Burnett, 1978; Cameron and Soost 1969; Esen and Soost, 1972; Oiyama, 1991; Starrantino and Recupero, 1982). The most recent evidence of triploid citrus varieties that were obtained by interploid sexual hybridization are the varieties of grapefruit (C. maxima (Burm) Merrill), Oro Blanco and Melogold that were developed in California (Soost and Cameron 1980, 1985). Like most citrus fruits, the Mexican lemon is diploid (2x). By crossing this species through sexual hybridization with tetraploid genotypes (2n= 4x) that contain characteristics of tolerance to the aforementioned health problems, it is possible to generate triploid varieties without seeds that tolerate these problems.
The conventional improvement of citrus has important limitations due to the complex reproductive biology of these species. Most genotypes are apomictic and develop adventitious embryos directly from the nucellular cells that limit or exclude the development of zygotic embryos (Kobayashi et al., 1979; Soost and Roose, 1996; Ruiz et al., 2000; Yildis et al., 2013). They contain the same genetic material as the mother plant, and can cause most polyembryonic cultivars to produce few hybrid plants. On the other hand, it is difficult to distinguish nucellar and zygotic plants at an early stage of development (De Lange and Vincent, 1977). So the identification of sexual embryos is important and require analysis such as cytology, flow cytometry, isoenzyme, or molecular analysis (Tusa et al., 2002).
According to Yildiz et al. (2013) many studies aimed at the separation of different types and varieties of citrus and the identification of nucellar and zygotic plants, began using morphological characterization (Furr and Reece, 1946; Cameron, 1979), spectrophotometry (Pieringer and Edwards, 1967) and chromatographic techniques (Albach and Redman, 1969; Stanley and Jurd, 1971; Tatum et al., 1974; Weinbaum et al., 1982). Several biochemical methods were also used, including enzymatic darkening (Esen and Soost, 1974).
None of these methods efficiently confirmed the identity of the true nucellar seedlings (Ruiz et al., 2000; Tusa et al., 2002). Later, isoenzymes were used (Iglesias et al., 1974; Moore and Castle, 1988; Ashari et al., 1988; Anderson et al., 1991). However, since the products of gene expression were used in these cases, the results could be influenced by the environment or by the stage of development of the plant and its organs, so this method was unreliable for the identification of zygotic seedlings.
The use of DNA polymorphisms for the identification of hybrid plants is important in citrus and other woody breeding programs, as this accelerates the progeny detection process. A series of recent studies have described the use of SSR markers as an alternative method to distinguish nucellar sexual seedlings in citrus fruits (Mullis et al., 1986; Ruiz et al., 2000; Oliveira et al., 2002; Rao et al., 2008; Shareefa et al., 2009; Yildis, et al., 2013).
This study describes the identification of Mexican lemon hybrids based on the molecular analysis of SSR markers.
Materials and methods
Biological material
For the study was used, a population of 203 plants originating from 13 Mexican lemon crosses with Italian lemons, citranges and somatic hybrids between citranges and sweet orange (Table 1), carried out from 2009 to 2012 in the Tecoman Experimental Field of INIFAP.
Table 1 List of hybrid Mexican lemon progenies used in the study.
| Crossing number | Crosses | Number of samples | Year of cross |
| 1 | Rosenberg (2x) X Colimex (2x) | 20 | 2009 |
| 2 | Colimex (2x) X Rosenberg (2x) | 14 | 2009 |
| 3 | Mex13 (4x) X Limoneira 8a (2x) | 12 | 2010 |
| 4 | Mex13 (4x) X Rosenberg (2x) | 19 | 2010 |
| 5 | Mex13 (4x) X Eureka (2x) | 10 | 2010 |
| 6 | Rosenberg (2x) X Mex20 (4x) | 8 | 2010 |
| 7 | Rosenberg (2x) X Mex13 (4x) | 7 | 2010 |
| 8 | Mex13 (4x) X C-35 (2x) | 50 | 2011 |
| 9 | Colimex (2x) X HS11 (4x) | 21 | 2011 |
| 10 | Colimex (2x) X Limequat (2x) | 6 | 2012 |
| 11 | Colimex (2x) X C-32 (2x) | 17 | 2012 |
| 12 | Colimex (2x) X C-swingle (2x) | 8 | 2012 |
| 13 | Colimex (2x) X Yuma (2x) | 11 | 2012 |
Extraction of genomic DNA
The total DNA was extracted from young leaves according to the method of Doyle and Doyle, 1987, with modifications Hernández-Nava (2013). In an eppendorf tube, 0.1 g of plant tissue was deposited together with 600 μl of saline buffer and crushed in a Retsch® MM400 disruptor at 25 frequencies (f/s) for three min. Subsequently, 2% CTAB extraction (1.4 M NaCl, 100 mM Tris, 20 mM EDTA, pH 8) and 3 μl of β-Mercaptoethanol at 100% were resuspended in extraction buffer. The contents of the vortex tube were mixed for 10 seconds and incubated at 55 °C for 30 min.
Next, 400 μl of phenol: chloroform: alcoholisoamyl was added in a ratio of 25:24:1 respectively, mixing the sample in vortex for 10 seconds. The sample was centrifuged for 10 min at 14 000 rpm at 4 °C and 500 μl of the aqueous phase was collected preventing the phases from mixing. Subsequently, 50 μl of ammonium acetate and 500 μl of isopropanol were added and the sample was incubated for 10 min at -20 °C. It was then centrifuged for 10 min at 14 000 rpm at 4 °C and the supernatant was decanted without losing the tablet that formed. 1 ml of 70% ethanol was added and centrifuged for 1 min at 14 000 rpm at 4 °C. After the time elapsed, the sample was dried at room temperature until the ethanol evaporated. Finally, the tablet was resuspended in 50 μl of RNase-free water and the sample was stored at -20 °C. The quality and purity of DNA obtained was quantified in Nanodrop 2000.
Amplification by PCR
The PCR amplification of specific segments of DNA by oligo sets reported by Kijas et al. (1997) (Table 2), was performed in a total volume of 20 μl using 10 μl of iTaq™ Universal Probes supermix (dNTPs, iTaq DNA polymerase, MgCl2, enhancers, stabilizers, dyes and ROX normalization) and 2.5 μl of each primer Forward and Reverse. The amplification program consisted of a cycle at 94 °C for 5 min, followed by 32 cycles of 94 °C for 1 min, 55 °C for 30 s and 72 °C for 1 min. A final extension of 4 min at 72 ºC. The hybridization temperature was reduced to 45 °C for 30 s to promote primer amplification TAA52.
Table 2 Microsatellite initiators used in the detection of lemon hybrids.
| Name | Sequence 5’-3’ | Sequence 3’-5’ |
| TAA41 | AGGTCTACATTGGCATTGTC | ACATGCAGTGCTATAATGAATG |
| TAA45 | GCACCTTTTATACCTGACTCGG | TTCAGCATTTGAGTTGGTTACG |
| TAA52 | GATCTTGACTGAACTTAAAG | ATGTATTGTGTTGATAACG |
| cAGG9 | AATGCTGAAGATAATCCGCG | TGCCTTGCTCTCCACTCC |
The amplification of the DNA fragments was carried out in a Bio-Rad T100™ Thermal Cycler. thermalcycler. The PCR amplification products were analyzed in 8% acrylamide gel and developed with 0.2% silver nitrate. A 1kb molecular weight marker (invitrogen) was used. The running conditions of the samples were 190 V and 200 mA for 90 min.
Analysis of data
The dendograms were made from the identification of presence (1) or absence (0) of bands using the four primers, through the unweighted paired grouping method with arithmetic means (UPGMA) and squared Euclidean distance. The analyzes were carried out with the use of the statistical package Numerical Taxonomy and Multivariate Analysis System (NTSYS-pc 2.21) (Rohlf, 2000).
Results and discussion
The hybrid plants were determined by the presence of male parent bands amplified by PCR using four primers. In Figure 1a and 1b, some of the hybrids identified with the primers cAGG9 and TAA45 are shown in one of the crosses evaluated, (Mex13 (4x) x Rosenberg (2x)). From 203 plants obtained from the different crosses, the best initiator was able to identify 145 (71%) zygotic plants. The results obtained are similar to those obtained by some authors. Frost and Soost (1967) reported zygotic frequencies of 78.7% and 14% in King and Willowleaf tangerines respectively, using P. trifoliata as male progenitor.
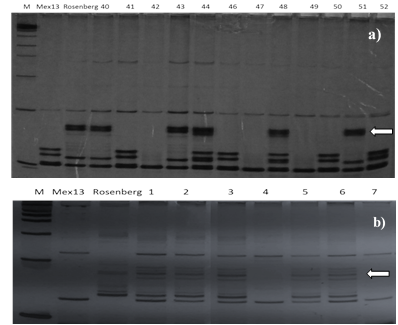
Figure 1 Polyacrylamide gel showing the SSR analysis of progeny obtained from Mexican lemon crosses. (a) Initiator cAGG9, (b) initiator TAA45 marker 1kb (M), female parent Mex13, male Rosenberg.
Other authors such as Hearn (1973) found that the ‛Mediterranean’ sweet orange selection produced 62% of zygotic seedlings when P. trifoliata was used as a male parent. In other reports (Soost et al., 1980) they found a zygotic frequency of approximately 85% when they used as a female parent the mandarin King and male to ‛Parson Special’. Several studies have shown that the frequency of zygotic seedlings does not exceed 15%, but this will depend on the species and in some cases only nucellar individuals are obtained (Cameron and Soost, 1980; Hirai et al., 1986; Ashari et al., 1988; Roose and Traugh, 1988).
Several levels of zygote seedlings have been observed in other citrus species and crosses. Some other authors, among them. Hwang and Yeuh (1989) reported zygotic frequencies of 42.8% and 66.7% in tangerine crosses ‛Tankan’ x P. trifoliata. Hwang (1991) reported that in seedlings identified by leaf morphology, he obtained 73% zygotic lemon seedlings ‛Eureka’ in crosses with trifoliate orange and 38% to 56% in ‛Eureka’ and ‛Lisbon’ lemon cultivars in crosses with sweet orange. In other works, such as that of Moore and Castillo (1988), only 24% of zygotic ‛Volkameriana’ lemon seedlings were identified; these results were based on isoenzyme-based markers.
With the use of SSR markers, Ruiz et al. (2000), classified 86.95% of hybrid seedlings of crosses between ‛Flying Dragon’ and tangor ‛Ortanique’. However, Yildiz et al. (2013), identified 86 seedlings (36. 91%) nucelar and 63% zygotic in crosses with mandarin ‛Fremont’ as female parent and the Rio Red variety as male parent. These same authors point out that when they used the mandarin ‛Robinson’ as the female parent, this proportion was 31.09% nucellar and 69% zygotic seedlings.
Data that differ from those obtained in this study were reported by Yun et al. (2007), who obtained 0% to 13.4% hybrid seedlings in crosses where the mandarins Miyagawa Wase, Okitsu Wase and Shiranuhi were used as female parents and Ponkan mandarin and Swingle citrumelo as male parents. Other authors such as Rao et al. (2008) crossed different tangerine and grapefruit cultivars to obtain alternative rootstocks to sour orange and with SSR markers they identified 22.2% of hybrid seedlings of these crosses.
According to García et al. (1999) zygotic embryos produced by self-pollination are less vigorous and may not be competitive with the nucellar. The percentages of zygotic progeny found in several citrus hybrids depend on the seed of the parent used (Spiegel-Roy et al., 1977), the origin of the pollen (Cameron and Soost, 1980; Soares-Filho et al., 1995) and environmental influences (Khan and Roose, 1988; Moore and Castle, 1988; Roose and Traugh, 1988).
The diploid, triploid and tetraploid plants of each combination were evaluated using SSR primers. The four managed to identify hybrid plants (Table 3). With the TAA45 primer it was possible to identify the largest number of hybrids (145), this being the best of all, followed by cAGG9 and TAA52 with 130 and 125 hybrids, respectively. In contrast to the results obtained in this study, they were reported by Yildiz et al. (2013) in their investigations located in a study population that the primers AG14 and TAA03 were more effective to distinguish zygotic individuals and the TAA45 primer was not able to identify the zygotic seedlings.
Table 3 Hybrid plants identified with four of the molecular markers reported by Kijas et al. (1997).
| Crosses | Initiator cAGG9 | Initiator TAA41 | Initiator TAA45 | Initiator TAA52 |
| Colimex X Yuma | 16 | 8 | 7 | 5 |
| Colimex X C-swingle | 2 | 9 | 2 | 2 |
| Colimex X Híbrido somático 11 | 25 | 20 | 20 | 14 |
| Colimex X Limequat | 5 | 0 | 2 | 2 |
| Colimex X Rosenberg | 7 | 8 | 11 | 12 |
| Colimex X C-32 | 3 | 2 | 16 | 2 |
| Mex13 X Eureka | 4 | 5 | 7 | 9 |
| Mex13 X Limoneira 8a | 7 | 8 | 10 | 3 |
| Mex13 X Rosenberg | 12 | 15 | 13 | 17 |
| Mex13 X C-35 | 39 | 32 | 48 | 53 |
| Rosenberg X Colimex | 7 | 6 | 3 | 2 |
| Rosenberg X Mex13 | 1 | 1 | 0 | 1 |
| Rosenberg X Mex20 | 2 | 0 | 6 | 3 |
| Total de plantas híbridas | 130 | 114 | 145 | 125 |
With respect to the TAA41 primer, it was this one that identified the least plants (114). Similar results were found by Yildiz et al. (2013), reported that in combinations where they used the Fremont tangerine as a female parent, the TAA41 initiator was able to identify only 47 zygotic plants that the rest of the SSR primers used. These same authors report that, in combinations where the ‛Robinson’ tangerine was used as the female parent, the AG14 and TAA03 primers identified 59 and 58 zygotic plants respectively, more than the TAA41 primer. In contrast, the primers CAT01, TAA01, TAA45 and TAA52 (when the mandarin ‛Fremont’ and ‛Robinson’ were used as female progenitors) were not useful in determining the identity of zygotic plants.
In populations that are derived from a cross, all individuals that are zygotic show a genotype different from that of the mother in any locus of discrimination, this will occur as long as the father has alleles different from those of the mother (Ruiz et al., 2000). The variation in the number of bands amplified by different primers may be due to factors such as the structure of the primer, the amount of template and the number of sites recognized in the genome (Muralidharan and Wakeland, 1993).
Other authors, including Nageswara et al. (2008) identified 77.8% nucellar seedlings and 22.2% zygotic seedlings, with EST-SSR markers, in introgression hybrids of mandarin (C. reticulata Blanco) and grapefruit (C. maxima Merr.). Other authors such as Mei-lian et al. (2007) reported in crosses of red tangerine (C. reticulata Blanco) X trifoliate orange (P. trifoliata (L.) 111 hybrid individuals through embryo rescue and verified by SSR markers and 44 hybrid plants from tangerine crosses Satsuma (C. unshiu Marc) X trifoliate orange (P. trifoliata L.).
In the same way Oliveira et al. (2002), report the identification of hybrids derived from crosses between tangor ‛Murcott’ [C. reticulata Blanco × C. sinensis (L.) Osb.] and sweet orange ‛Pera’. [C. sinensis (L.) Osb.], with analysis of SSR markers. Siragusa, et al., 2006, identified and evaluated the genetic variability of sweet orange accessions with molecular markers SSR. Also Aleza et al., 2012, identified hybrids that show the specific allele of the male parent with molecular markers SSR in crosses 4x X 2x of several citrus fruits. Aleza et al. (2010) used SSR markers to show that all plants recovered from undeveloped seeds obtained from 2x X 4x sexual hybridizations were the result of the zygotic embryo.
In citrus fruits, SSR markers can easily distinguish between species or varieties that arise from sexual hybridization (Barkley et al., 2006; Luro et al., 2008). On the contrary, the differentiation of genotypes derived from spontaneous mutations is very difficult, as in the case of clementines (Breto et al., 2003) or sweet oranges (Fang and Roose, 1997). By using several heterozygous markers differentiating male and female parents, one can also find a good chance of differentiating between independent zygotic plants.
Fang and Roose (1997) mention that mutations in most cases can cause significant changes in tree morphology, but with modifications that occur in the DNA sequence, they are difficult to detect. Novelli et al. (2000, 2006) also reported that despite considerable morphological differences, there are few RFLP, RAPD and SSR polymorphisms. On the basis of SSR markers, Cao et al. (2007) could not detect differences between 16 Satsuma tangerines and therefore the possible origin of the mutation of these variants.
Seedlings always have a higher similarity value with their female parents, thereby indicating the decrease in genetic background, the increase in inbreeding may be the cause of low tree yield in various environmental conditions (Fehr, 1993).
When carrying out the dendograms with the data of the four SSR markers of the individuals obtained with the different crosses, it was possible to detect that the coefficient of similarity of the Euclidean distance varied from 0 to 999.09 among the 203 individuals that were analyzed. This is due to the great variability generated in these crosses, since the genomes of different species and even of different genera were combined.
On the other hand, both the Mexican lemons and the Italian lemons are in heterozygous form and therefore by segregating the alleles in the gametes and then by recombining in the crossing, they are generating new genetic combinations. On the other hand, the citranges are formed by the genome of C. sinesis and P. trifoliata, also in highly heterosigotic condition, which increases the genetic diversity. This result coincides with that reported by Herrero et al. (1996) who point out that limes and lemons show a high percentage of heterozygosity
Figure 2 shows one of the 18 dendograms constructed with the SSR profiles obtained in the cluster analysis (UPGMA). The dendrogram of the Mex13 x Rosenberg crosses was separated into four groups. The first was formed by the female parent Mex13. In the second are the individuals H40, H43, H44, H48, H51, H41, H42, H55, H53, H54, H50, H52, H46, H47, H49 and the male parent Rosenberg. The third group was formed by the individual H45. And in the last are the indices H196, H209 and H198. In this cross it was observed that all the individuals analyzed were identified as hybrids. The individuals H40, H43, H44, H48 and H51 are genetically similar to Rosenberg, which indicates that the four initiators were able to identify regions similar to the male parent.
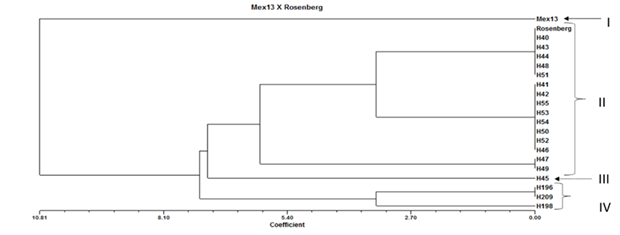
Figure 2 Dendogram generated with data from four SSR markers of individuals obtained from the cross of ‛Mex13’ x ‛Rosenberg’ constructed with the UPGMA method.
The rest of the hybrids were identified by three of the primers used. In a study of citrus germplasm, Shahsavar et al. (2007) reported that lemons (C. limon L. Burm F.), Lisbon, ‛Lemon Rock’ and ‛pear shape limon’ were separated from other genotypes when analyzing the polymorphism with molecular markers ISSR with similarity value of 0.65. In this same work these authors report that the populations of plants studied exhibit very similar morphological characteristics; however, molecular phenotypes revealed substantial differences in similarity between hybrids and progenitors.
The SSR are codominant and multialelic markers that usually show high levels of polymorphism and that have a high degree of reliability and reproducibility (Morgante and Olivieri, 1993). These markers are the most powerful to understand the detailed patterns of kinship composition and individual discrimination in clones, so they are widely used for the identification of cultivars and genetic maps (Tautz, 1989, Nassar and Collevatti, 2005). When a population is derived from a cross, all zygotic individuals show a genotype different from that of the mother at any locus of discrimination, as long as the father has alleles different from those of the mother (Ruiz et al., 2000). In the case of a cross between homozygous or closely related parents, codominant markers can unequivocally discriminate the zygotic offspring of nucelar (Nageswara et al., 2008).
Conclusions
Hybrid Mexican lemon plants were identified with Italian lemons and somatic hybrids between citranges and sweet orange.
The four initiators allowed the identification of hybrid plants. The greatest number of hybrid plants were identified with the primers TAA45 and cAGG9.
The identification of the hybrids of the population under study through the analysis of molecular markers, will save time and reduce the costs of maintenance of field material in breeding programs that involve obtaining a large number of seedlings derived from the crosses made.
The results of this study suggest that molecular marker analysis can contribute significantly to citrus breeding programs.
Literatura citada
Albach, R. F. and Redman, G. H. 1969.Composition and inheritance of flavanones in citrus fruit. Phytochemistry 8:127-143. [ Links ]
Aleza, P.; Juarez, J.; Cuenca, J.; Ollitrault, P. and Navarro, L. 2010. Recovery of citrus triploid hybrids by embryo rescue and flow cytometry from 2x 3 2x sexual hybridisation and its application to extensive breeding programs. Plant Cell Rep. 29:1023-1034. [ Links ]
Aleza, P.; Juárez, J.; Hernández, M.; Ollitrault, P. and Navarro, L. 2012. Implementation of extensive citrus triploid breeding programs based on 4x×2x sexual hybridisations. Tree Genetics and Genomes. 8:1293-1306. [ Links ]
Anderson, C. M.; Castle, W. S. and Moore, G. A. 1991. Isozymic identification of zygotic seedlings in Swingle citrumelo Citrus paradise and Poncirus trifoliata nursery and field populations. J. Am. Soc. Hort. Sci. 116:322-236. [ Links ]
Ashari, S.; Aspinall, D. and Sedgley, M. 1988. Discrimination of zygotic and nucellar seedlings of five polyembryonic citrus rootstocks by isozyme analysis and seedling morphology. J. Hort. Sci. 63:695-703. [ Links ]
Barkley, N. A.; Roose, M. L.; Krueger, R. R. and Federici, C. T. 2006. Assessing genetic diversity and population structure in a citrus germplasm collection utilizing simple sequence repeat markers (SSRs). Theor. Appl. Genet. 112:1519-1531. [ Links ]
Breto’, M. P.; Ruiz, C.; Pina, J. A. and Asíns, M. J. 2001. The diversification of Citrus clementina Hort. ex Tan., a vegetatively propagated crop species. Mol Phylogenet E. 21:285-293. [ Links ]
Cameron, J. W. 1979. Sexual and nucellar embryony in F1 hybrids and advanced crosses of Citrus and Poncirus. J. Am. Soc. Hort. Sci. 104:408-410. [ Links ]
Cameron, J. W. and Soost, R. K. 1980. Mono and poly embryony among tetraploid Citrus hybrids. HortSci. 15:730-731. [ Links ]
Cameron, J. W. and Burnett, R. H. 1978. Use of sexual tetraploids seed parent for production of triploids citrus hybrids. HortScience. 13:167-169. [ Links ]
Cameron, J. W. and Soost, R. K. 1969. Characters of new populations of citrus polyploids and the relation between tetraploidy in the pollen parent and hybrid tetraploid progeny. Proc. First int. Citrus Symposium. 1: 199-205. [ Links ]
Cao, Q. Q.; Meng, H. J.; Wen, X. P.; Yi, H. L. and Deng, X. X. 2007. Genetic diversity of male sterile and low fertility germplasm of Citrus revealed using SSR markers. Chin. J. Agr. Biotec. 4:99-104. [ Links ]
Cristofani, M. and Machado, M. A. 1998. Utilização de marcadores moleculares na identificação de plântulas zigóticas e nucelares em sementeira de limão ‘Cravo’. Laranja 19:147-158. [ Links ]
Cristofani, M.; Machado, M. A.; Targon, M. P. L. N. and Takita, M. A. 2002. Tristeza tolerance by improvement methods. In: 7th International citrus seminar: improvement. (Eds.). Donadio, L. C. y Sanches-Stuchi, E. Citrus Experimental station of Bevedoro, SF Brazil. 71-73 pp. [ Links ]
De Lange, J. H. and Vincent, A. P. 1977. Citrus breeding: new techniques in stimulation of hybrid production and identification of zygotic embryos and seedlings. Second Meeting of the International Society of Citriculture. Florida, USA. 2:589-595. [ Links ]
Doyle, J. J. and Doyle, J. L. 1987. Arapid DNA isolation procedure from small quantities of fresh leaf tissue. Phytochem. Bull. 19:11-15. [ Links ]
Esen, A. and Soost, R. K. 1974. Inherence of browning of young-shoot extracts of Citrus. J. Hered. 65:97-100. [ Links ]
Esen, A. and Soost, R. K. 1972. Tetraploids progenies from 2x x 4x crosses in citrus and their origin. J. Amer. Soc. Hort. Sci. 97:410-414. [ Links ]
Fang, D. Q. and Roose, M. L. 1997 Identification of closely related Citrus cultivars with inter-simple sequence repeat marker. Theor. Appl. Genet. 95:408-417. [ Links ]
Fehr, W. R. 1993. Principles of cultivar development. Macmillian Publishing Company. USA. [ Links ]
Frost, H. B. and Soost, R. K. 1967. Seed reproduction: development of gametes and embryos. In: Reuther, W.; Batchelor, L. D.; Webber, H. J. (Eds.). The Citrus industry. University of California Press, Berkeley, USA. 290-324 p. [ Links ]
Furr, J. R. and Reece, P. C. 1946. Identification of hybrid and nucellar citrus seedlings by a modification of the rootstock color test. J. Am. Soc. Hort. Sci. 48:141-146. [ Links ]
Garcia, R.; Asin, M. J.; Forner, J. and Carbonell, E. A. 1999. Genetic analysis of apomixes in Citrus and Poncirus by molecular markers. Theor. Appl. Genet. 99:511-518. [ Links ]
Grosser, J. W.; Ollitrault, P. and Olivares, F. O. 2000. Somatic hybridization in citrus: an effective tool to facilitate variety improvement. In Vitro Cell Dev. Biol. 36:434-449. [ Links ]
Grosser, J. W. and Gmitter, F. G. 2005. SIVB Congress Symposium Proceedings “Thinking outside the cell”: applications of somatic hybridization and cybridization in crop improvement, with citrus as a model. In vitro cell Dev. Biol. Plant 41: 220-225. [ Links ]
Hearn, C. J. 1973. Recently released cultivars and the development of new hybrids of Citrus fruits in Florida. First Meeting of the International Society of Citriculture, Murcia-Spain. 81-85 pp. [ Links ]
Hernandez, N. G. A. 2013. Prevalencia de Toxoptera citricida y tasa de adquisición del Citrus tristeza virus en la Península de Yucatán. Tesis de Maestria. Montecillo, Texcoco, Estado de México. 83 p. [ Links ]
Herrero, R.; Asins, M. J.; Carbonell, E. A. and Navarro, L. 1996. Genetic diversity in the orange subfamily Aurantioideae. I. Intraspecies and intragenus genetic variability. Theor. Appl. Genet. 92:599-609. [ Links ]
Hirai, M.; Kozaki, I. and Kajiura, I. 1986. The rate of spontaneous inbreeding of trifoliate orange and some characteristics of the inbred seedlings. Jpn. J. Breed. 36:138-146. [ Links ]
Hwang, A. S. 1991. The polyembryony and identification of zygotic seedlings of lemon. J. Agr. Res. China. 40:225-232. [ Links ]
Hwang, A. S. and Yeuh, C. S. 1989. Studies on polyembryony and improvement of breeding efficiency of oranges. Tai Agr. Res. Ins. Spe. Pub. 27:28-38. [ Links ]
Iglesias, L.; Lima, H. and Simon, J. P. 1974. Isozyme identification of zygotic and nucellar seedlings in Citrus. J. Hered. 65:81-84. [ Links ]
Iwamasa, M.; Nito, N. and Ling, J. T. 1988. Intra and inter generic hybridization in the orange subfamily Aurantioidea. Proc. Int. Soc. Citricult. 1:123-130. [ Links ]
Khan, I. A. and Roose, M. L. 1988. Frequency and characteristics of nucellar and zygotic seedlings in three cultivars of trifoliate orange. J. Amer. Soc. Hort. Sci. 113:105-110. [ Links ]
Kijas, J. M. H.; Thomas, M. R.; Fowler, J. C. S. and Roose, M. L. 1997. Integration of trinucleotide microsatellites into a linkage map of Citrus. Theor. Appl. Genet. 94:701-706. [ Links ]
Kobayashi, S.; Ieda, I. and Nakantani, M. 1979. Studies on the nucellar embryogenesis in Citrus II. Formation of the primordium cell of the nucellar embryo in the ovule of the flower bud, and its meristematic activity. J. Jpn. Soc. Hort. Sci. 48:179-185. [ Links ]
Luro, F.; Costantino, G.; Terol, J. F.; Argout, X.; Allario, T.; Wincker, P.; Talon, M.; Ollitrault, P.; and Morillon, R. 2008. Transferability of the EST-SSRs developed on Nules clementine (Citrus clementina). BMC Genomics. [ Links ]
Mei, L. T.; Jian, K. S. and Xiu, X. D. 2007 Production of two mandarin trifoliate orange hybrid. Euphytica, 157. 155-160 [ Links ]
Moore, G. A. and Castle, W. S. 1988. Morphological and is ozymic analysis of open-pollinated Citrus rootstock populations. J. Hered. 79:59-63. [ Links ]
Morgante, M. and Olivieri, A. M. 1993 PCR-amplified microsatellite as markers in plant genetics. Plant J 3:175-182. [ Links ]
Mullis, K.; Fallona, S.; Schraf, S.; Saiki, R.; Horn, G. and Erlich, H. 1986. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harbor Symp Quant Biol 51:263-273. [ Links ]
Muralidharan, K. and Wakeland, E. K. 1993. Concentration of primer and template qualitatively affects products in random-amplified polymorphic DNA PCR. Biotechniques 14:362-390. [ Links ]
Nageswara, M. R.; Soneji, J. R.; Chen, C.; Huang, S. and Gmitter, F. G. Jr. 2008. Characterization of zygotic and nucellar seedlings from sour orange-like citrus rootstock candidates using RAPD and EST-SSR markers Tree Genetics Gen. 4:113-124. [ Links ]
Nassar, N. M. A. and Collevatti, R. G. 2005 Microsatellite markers confirm high apomixis level in cassava bred clones. Heriditas. 142:33-37. [ Links ]
Navarro, L.; Olivares, F. O.; Juárez, J.; Aleza, P.; Piña, J.; A. Cerrera, M.; Fagoaga, C.; Pérez, R. M. and Peña, L. 2002. Citrus improvement in Spain through triploid breeding, somatic hybridization and genetic trsnformation. In: 7th International citrus seminar: Improvement. (Eds.). Donadio, L. C. y Sanches-Stuchi, E. Citrus Experimental station of Bevedoro, SF Brazil. 57-70 pp. [ Links ]
Novelli, V. M.; Cristofani, M.; Souza, A. A. and Machado, M. A. 2006. Development and characterization of polymorphic microsatellite markers for the sweet orange (citrus sinensis L. Osbeck). Gen. Mol. Biol. 29:90-96. [ Links ]
Novelli, V. M.; Machado, M. A. and Lopes, C. R. 2000. Isoenzymatic polymorphism in Citrus spp. and P. trifoliata (L.) Raf. (Rutaceae). Genetics and Molecular Biology 23:163-168. [ Links ]
Oiyama, I. 1991. Use of pollen from somatic hybrid between Citrus and Poncirus in the production of triploids. HortSci. 1082. [ Links ]
Oliveira, A. C.; Garcia, A. N.; Cristofani, M. and Machado, M. A. 2002. Identification of citrus hybrids through the combination of leaf apex morphology and SSR markers. Euphytica 128:397-403. [ Links ]
Ollitrault, P.; Dambier, D.; Vanel, F. and Froelicher, Y. 2000. Creation of triploid Citrus hybrids by electrofusion of haploid and diploid protoplasts. Proceedings of First International Citrus Biotechnology Symposium. Acta Hortic. 535:191-198. [ Links ]
Pieringer, A. P. and Edwards, G. J. 1967. Identification of nucellar and zygotic citrus seedlings by infrared spectroscopy. J. Am. Soc. Hort. Sci. 86:226-234. [ Links ]
Rao, M. N.; Soneji, J. R.; Chen, C.; Huang, S. and Gmitter, F. G. 2008. Characterization of zygotic and nucellar seedlings from sour orange-like Citrus rootstock candidates using RAPD and EST-SSR markers. Tree Gen. Genom. 4:113-124. [ Links ]
Rohlf, F. J. 2000. NTSYS-pc: numerical taxonomy and multivariate analysis system, ver. 2.10e. Exeter Ltd., Setauket. [ Links ]
Roose, M. L. and Traugh, S. N. 1988. Identification and performance of Citrus trees on nucellar and zygotic rootstocks. J Am. Soc. Hort. Sci. 113:100-105. [ Links ]
Ruiz, C.; Breto, M. P. and Asíns, M. J. 2000. A quick methodology to identify sexual seedlings in citrus breeding programs using SSR markers. Euphytica. 112:89-94. [ Links ]
Shahsavar A. R.; Izadpanah, K.; Tafazoli, E. and Sayed, T. B. E. 2007. Characterization of citrus germplasm including unknown variants by inter-simple sequence repeat (ISSR) markers. Sci. Hortic. 112:310-314. [ Links ]
Shareefa, M.; Singh, A. K.; Manish, S. and Dubey, A. K. 2009. Differentiation of nucellar and zygotic seedlings in citrus using ISSR markers. Ind. J. Agr. Sci. 79:884-889. [ Links ]
Shi, P. Z.; Jian, K. S.; Zhi, Y. H.; TAN, B.; Zong, Z. X.; Hua, L. Y.; Xiu, S. S. and Rajam, V. M. 2009. Citrus biotechnology: achievements, limitations and future directions. Physiol. Mol. Biol. Plants. 15(1). [ Links ]
Siragusa, M.; Depascuale, F.; Abbate, L. and Tusa, N. 2006. Identification of sour orange accessions and evaluation of their genetic variability by molecular marker analyses. HortSci. 41(1):84-89. [ Links ]
Soares, F. W. dos S.; Lee, L. M. and da Cunha, S. A. P. 1995. Influence of pollinators on polyembryony in Citrus. Acta Hortic. 403:256-265. [ Links ]
Soost, R. K.; Williams, T. E. and Torres, A. M. 1980. Identification of nucellar and zygotic seedlings of Citrus with leaf isozymes. HortSci. 15:728-729. [ Links ]
Soost, R. K. and Cameron, J. W. 1980. ‛Oroblanco’, a triploid pummelo grapefruit hybrid. HortScience. 15:667-669. [ Links ]
Soost, R. K. and Cameron, J. W. 1985. ‘Melogold’ a triploid pummelo-grapefruit hybrid. HortScience 20:1134-1135. [ Links ]
Soost, R. K. and Roose, M. L. 1996. Citrus, chapter 6. In: Janick, J. and Moore, J. N. (Eds.). Fruit breeding: tree and tropical fruits. Wiley, New York. 257-323 pp. [ Links ]
Spiegel, R. P.; Bardi, A. and Shani, A. 1977. Peroxidase isozymes as a tool for early separation of nucellar and zygotic citrus seedlings. In: Second Meeting of the International Society of Citriculture. Florida-USA. 2:619-624. [ Links ]
Stanley, W. L. and Jurd, L. 1971. Citrus coumarins. J. Agr. Food Chem. 19:1106-1110. [ Links ]
Starrantino, A. and Recupero, G. 1982. Citrus hybrids obtained in vitro from 2x females by 4x males. Proc. Int. Soc. Citricult. 1:31-32. [ Links ]
Tatum, J. H.; Berry, R. E. and Hearn, C. J. 1974. Characterization of Citrus cultivars and separation of nucellar and zygotic seedlings by thin layer chromatography. Proc. Florida State Hort. Soc. 87:75-81. [ Links ]
Tautz D. 1989. Hypervariability of simple sequences as a general source of polymorphic DNA markers. Nucl Acid Res 17:6463-6471. [ Links ]
Tusa, N.; Abbate, L.; Ferrante, S.; Lucretti, S. and Scarano, M. T. 2002. Identification of zygotic and nucellar seedlings in Citrus interploid crosses by means of isozymes, flow cytometry and ISSR-PCR. Cel and Mol Bio Letters 7:703-708. [ Links ]
Tusa, N.; Fatta del Bosco, S.; Nardi, L. and Lucretti, S. 1996. Obtaining triploid plants by crossing citrus lemon cv. ‘Femminello’ 2N x 4N allotetraploid somatic hybrids. Proc. Int. Soc. Citricult. 1:133-136. [ Links ]
Weinbaum, S. A; Cohen, E. and Spiegel, R. P. 1982. Rapid screening of ‘Satsuma’ mandarin progeny to distinguish nucellar and zygotic seedlings. Hort. Sci. 17:239-240. [ Links ]
Yildiz, E.; Kaplankiran, M.; Demirkeser T.; Uzun, H. A. and Toplu, C. 2013. Identification of Zygotic and Nucellar Individuals Produced from Several Citrus Crosses Using SSRs Markers. Not. Bot. Hort. Agrobo. 41(2):478-484. [ Links ]
Yun, J. U.; Yang, H. B.; Jung, Y. H.; Yun, S. H.; Kim, K. S.; Kim, C. S. and Song, K. J. 2007. Identification of zygotic and nucellar mandarin seedlings using randomly amplified polymorphic DNA. Hort. Environ. Bio. 48:171-175. [ Links ]
Received: December 00, 2017; Accepted: January 00, 2018











 texto em
texto em 


