Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Revista mexicana de ciencias agrícolas
versión impresa ISSN 2007-0934
Rev. Mex. Cienc. Agríc vol.8 no.8 Texcoco nov./dic. 2017
https://doi.org/10.29312/remexca.v8i8.708
Essays
Isolation and diagnosis protocols of Phytophthora spp. applied research approach
1Universidad Michoacana de San Nicolás de Hidalgo-Instituto de Investigaciones Agropecuarias y Forestales. Carretera Morelia-Zinapécuaro-Tarímbaro km 9.5, Michoacán, México. CP. 58880. 01 (443) 1940975. (alexsotoppv@gmail.com; gra.labpv@gmail.com; marle-dc@yahoo.com.mx).
2Colegio de Posgraduados-Programa de Fruticultura. Carretera México-Texcoco km 36.5, Montecillo, Texcoco, Estado de México. México. CP. 56230. Tel. 01 (55) 24905471.(fernandezpavia@hotmail.com;yletif@yahoo.com.mx;lexquilax@yahoo.com.mx).
3Facultad de Agrobiología-Universidad Michoacana de San Nicolás de Hidalgo. Paseo Lázaro Cárdenas esq. Berlín, Colonia Viveros, Uruapan, Michoacán, México. CP. 60090. 01 (452) 1126710. (marelpesa@yahoo.com.mx).
The purpose of this review is to make known to the researcher, fundamental protocols that are used for the isolation, diagnosis and identification of Phytophthora spp., covering general aspects of the genus for a better understanding and analysis of the same. This information will be useful for researchers working with diseases caused by this oomycete, and is mainly addressed to countries in Latin America. Currently, conventional and molecular methods are used for the detection and identification of Phytophthora species. Conventional methods include the use of taxonomic keys, observation of symptoms and detection of signs of the pathogen in diseased plants, and isolation of the causative agent of infected plant tissue, soil surrounding roots and water sources used for irrigation, using general or selective culture media. Molecular methods provide results that are more accurate in Phytophthora identification and can be performed in shorter time periods and it is not essential that the researcher has experience in aspects of the morphology of members of this genus. It is advisable to integrate the morphological and molecular characterization for the identification, description and taxonomy of Phytophthora.
Keywords: Phytophthora; isolated; medium selective
El propósito de esta revisión es dar a conocer al investigador, protocolos fundamentales que se emplean para el aislamiento, diagnóstico e identificación de Phytophthora spp., cubriendo aspectos generales del género para una mejor comprensión y análisis del mismo. Esta información será de utilidad para investigadores que trabajan con enfermedades causadas por este oomicete, y va dirigida a países de América Latina. Actualmente, se emplean métodos convencionales y moleculares para la detección e identificación de especies de Phytophthora. Los métodos convencionales incluyen el uso de claves taxonómicas, la observación de síntomas y detección de signos del patógeno en las plantas enfermas y el aislamiento del agente causal del tejido vegetal infectado, del suelo que rodea a las raíces y de las fuentes de agua utilizadas para el riego, usando medios de cultivo generales o selectivos. Los métodos moleculares proporcionan resultados que son más precisos en la identificación de Phytophthora, además de que se pueden realizar en lapsos de tiempo más cortos y no es indispensable que el investigador tenga experiencia en aspectos de la morfología de los miembros de este género. Es recomendable integrar la caracterización morfológica y molecular para la identificación, descripción y taxonomía de Phytophthora.
Palabras clave: Phytophthora; aislados; medio selectivo
The species of the genus Phytophthora are hemibiotrophic pathogens of great importance in agricultural and forestry crops as they cause great economic losses in a wide range of plant species in the world (Erwin and Ribeiro, 1996; Hall and Albers, 2009; Jung et al., 2000; Roy and Grünwald, 2014). Among the species of this oomycete that cause greater damage or loss are: Phytophthora ramorum, causal agent of the disease called sudden oak death, which also affects other trees and shrubs, P. palmivora causing bud rot, buds and fruits in palms; P. capsici pathogen that causes wilting and rotting of crown and root in chile, cucurbits and other commercial crops, P. infestans causing late blight of potato, tomato and other plants of the family Solanaceae, P. cinnamomi that infects More than 3,000 species of plants in the world (Erwin and Ribeiro, 1996; Cooke et al., 2000; Drenth and Guest 2004; Shearer et al., 2004; Hardham, 2005; Dart and Chastagner, 2007; Sutton et al., 2007; Brasier, 2008; Hansen et al., 2012; Jiang and Tyler, 2012; Kroon et al., 2012; Lamour et al., 2012).
This last species is an example of the economic and ecological impact that some species of Phytophthora can have in the world. For example, 2 284 (40%) of the 5 710 plant species described in southwestern Australia have been reported to be susceptible to P. cinnamomi. This pathogen cost the Australian economy 1.6 billion dollars over the course of a decade (Carter, 2004).
The number of species described in the genus Phytophthora has doubled in the last decade, and is likely to increase considerably as more studies are carried out on forests and water bodies. On the other hand, multigenic phylogenetic analyzes, molecular techniques, genomic sequencing and databases with Phytophthora sequences have made possible a more accurate identification and determination of the actual number of described species of this pathogen in the world. Currently 152 species belonging to the genus Phytophthora have been formally described http://idtools.org:8080/key_server/player.jsp?keyId=48. The Phytophthora species listed on the Internet are http://www.phytophthoradb.org, http://www.phytophthora-id.org, http://www.qbank.eu, and http://www.boldsystems.org.
Means of cultivation
The culture media selected to isolate, describe and identify Phytophthora species are of paramount importance. One factor that must be considered in choosing the appropriate medium is the Phytophthora species with which it is being worked (Erwin and Ribeiro, 1996). The selective means to isolate Phytophthora contain fungicides and antibiotics, to avoid the growth of fungi and bacteria.
The most commonly used selective media are PARPH, PARP, PARPH, PVP (P-pimaricina, A-ampicilina, R-rifampicina, P-pentacloronitrobenceno, H-himexazol, V-vancomicina), 3-P (P-pimaricina, P-Penicinila, P-Polimixina B, CARP (C-cornmeal agar, A-ampicilina, R-rifamicina, P-pimaricina), CARPD (D-dicloran), CARP+ (+-benlato e himexazol), there are variations in the concentrations of the antibiotics and fungicides are added (Tsao and Ocana, 1969; Jeffers and Martin, 1986; Erwin and Ribeiro, 1996; Drenth and Sendall, 2001) these must be added after the medium has been sterilized the medium. Pimaricin is an expensive antibiotic for laboratories in Latin America; however, it can be replaced by Delvocid Instant (Erwin and Ribeiro, 1996), which is inexpensive, it contains Natamycin, which is why this medium is known as NARPH. Likewise, the himexazole that previously came from Japan, is currently available in different countries. The selective medium can be prepared using corn flour agar or V8 agar. When corn flour can not be purchased, it can be replaced by corn starch (Felix- Gastelum. Comun. Pers., 2015).
Some examples in which the medium is quite specific is that of rye agar, which is mainly used to isolate P. infestans, P. ipomoeae and P. mirabilis, 3-P media and PCH (P-pimaricina, C-cloranfenicol, H-himexazol) are specific for isolating P. cinnamomi from roots and soil (Drenth and Sendall, 2001; Ivors, 2016). The protocols and media (V8 agar, liquid V8, super V8, V8-rye, rye A, rye B, oatmeal, 5% carrot agar, fresh lime and sucrose), currently employed are described in the Ivors (2016). In each laboratory, culture media can be prepared with the components available, it is also recommended to test different methods of preparation and select the most suitable ones for the individual needs.
Direct diagnosis
When a phytopathological diagnosis is carried out, the results are expected to be obtained as quickly as possible, for which kits, ELISA tests, fluorescence in situ hybridization (FISH), hyperspectral techniques, antibody based biosensors, immunoflorescence, fluorescence, thermography, and test strips for the direct detection of pathogens in the field that are of great help and can detect Phytophthora (Miller et al., 1994; Fang and Ramasamy, 2015).
Most field-use kits based on serological techniques give a rapid on-site response in a few minutes or hours, which is of great use to producers, consultants, technicians and inspectors. Tests can be performed from any plant tissue that is required to be analyzed and the result is easy to interpret. If necessary, the results can be confirmed later in the laboratory. Some of the kits available on the market are Phytophthora late blight Pocket Diagnostic® test for the detection of P. infestans in potato, Phytophthora spp. ALERT-LF® and ImmunoStrips to identify infections caused by Phytophthora in a wide range of crops including nursery plants. These kits are designed for use in field or laboratory from plant tissue samples, do not require special equipment to carry them out, since all the required material is included.
Isolation of Phytophthora
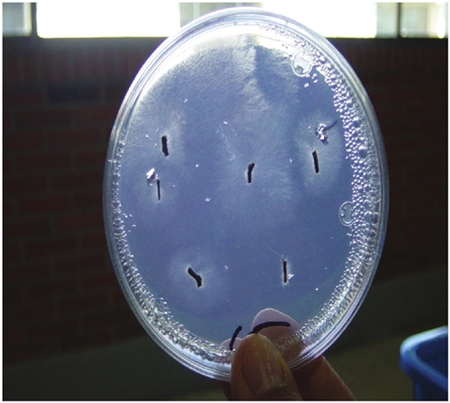
Isolation of Phytophthora from plant tissue. A suitable collection and selection of samples determine the success in the process of isolation of Phytophthora from diseased plant tissue. It is recommended to collect samples of leaves, stems, roots, fruits, flowers, etc, that have a zone of healthy tissue in front of the advancement of the lesion(Figure 1). To isolate sections of tissue from the area mentioned should be cut, as it will be where the pathogen is in active growth. These tissues can be planted without disinfesting, directly in a selective medium (Erwin and Ribeiro, 1996; Streito, 2002; Jung and Blaschke, 2004; Jung and Burgess, 2009; Kasuga et al., 2012).
It is not recommended to isolate Phytophthora from necrotic tissue due to the presence of fungal and saprophytic bacteria. Some authors suggest to disinfect the tissues superficially, for which the sections of tissue are immersed in 70% ethanol for 10 to 30 s, then washed with sterile distilled water, then dried with sterile filter paper and finally placed in selective culture medium (Hüberli, 2000; Davison and Tay, 2005; Ghimire et al., 2009; Moralejo et al., 2009; Tsopmbeng et al., 2012), in some reports it is mentioned that alcohol does not leave residues so it is not necessary to rinse (Martin et al., 2012; Borines et al., 2014).
A 10% commercial chlorine solution (5% sodium hypochlorite) is also used mainly. The disinfestation process involves immersing the plant tissue in the chlorine solution for 30 s, then rinsing with sterile distilled water and then drying with sterile paper, finally cutting the tissue sections before placing them in the selective medium (Drenth and Sendall, 2001; Ghimire et al., 2009).
The disinfestation time may vary, which depends on the type of tissue from which the insulation is to be performed. It is important to take into account that sensitivity to chlorine varies from one species to another of Phytophthora. Hong et al., (2003), have shown that the sensitivity to this is pathogen-dependent. Some species of Phytophthora may be sensitive to chlorine and ethanol and should be omitted (Hong et al., 2003). When it is unknown if the species is sensitive to these compounds, it is recommended to disinfect only some sections of tissue with ethanol or chlorine and others to seed directly in selective medium.
The tissue should preferably be dried with filter paper or sterile paper towels before being sown in any medium to prevent bacterial growth (Erwin and Ribeiro, 1996). Tissue sections seeded in selective medium should be incubated at a temperature favoring Phytophthora growth (Erwin and Ribeiro, 1996; Ferguson and Jeffers, 1999; Drenth and Sendall, 2001; Martin et al., 2012).
Isolation of Phytophthora of water. Irrigation water is an important source of inoculum of plant pathogens. The genera Phytophthora and Pythium of the family Pythiaceae are the most common, destructive and polluting in irrigation systems (Erwin and Ribeiro, 1996; Drenth and Sendall, 2001; Loyd et al., 2014; Zappia et al., 2014). More than 30 species of Phytophthora have been detected in water in the world, with some species not yet tested for pathogenicity (Zappia et al., 2014), others that have been inoculated in different hosts have not been pathogenic (Copes et al., 2015).
Only the pathogenicity of some P. irrigata in azalea, chili and tomato has been demonstrated; P. hydropathica in azalea, cineraria and tomato; P. megasperm in azalea and chili; P. nicotianae in vinca, begonia, eggplant, chili, cineraria, lupinus and tomato; P. palmivora in azalea, begonia, eggplant, chilli and tomato; P. tropicalis on azalea, begonia, eggplant, chili, cineraria, lupinus, tomato and vinca (Hong et al., 2008; Loyd et al., 2014). Water reservoirs or channels for agricultural use can be contaminated by water runoff from fields infested with Phytophthora, which may also lead to the dispersion of the pathogen to places where it was not present (Erwin and Ribeiro, 1996).
Recirculated water infested within and between nurseries favors the dispersal of Phytophthora propagules in the form of zoospores, healthy plants and areas where it was not found (MacDonald et al., 1994; Stewart-Wade, 2011). Therefore, effective protocols to detect this oomycete in irrigation water are essential. Filtration methods can process limited amounts of water compared to the enormous amount of water that exists in dams, bodies of water and irrigation systems.
In a study by Hong et al. (2002), obtained higher recovery rates in different water sources by filtering the water samples through a membrane that retains the propagules, which is then inverted and placed directly in the selective PARP-V8 medium. The membranes tested were of different materials and pore sizes, observing better results with the Millipore 5 and Durapore 5 membranes (Millipore®) obtaining higher recovery rates.
Isolation of soil Phytophthora or planting substrate
Phytophthora can be dispersed to new sites through the sale of different seed substrates or propagule-infested soils (Ferguson and Jeffers, 1999; Dart et al., 2007; Dart and Chastagner, 2007). It is recommended to use different trap vegetable tissues with each soil sample or substrates suspected to be contaminated with Phytophthora to rule out false negative results (Jeffers and Aldwinkle, 1987; Davison and Tay, 2005b; Balci et al., 2007; Tooley and Carras, 2011). A wide range of trap plant tissues are used to recover Phytophthora from soil since not all are colonized in the same way by different Phytophthora species (Erwin and Ribeiro, 1996). In a later paragraph the recommended trap vegetable tissues are mentioned.
To detect Phytophthora in soils or substrates, flooding of these with sterile distilled water is used, in such a way that propagules, especially zoospores are released into the water, swim to the surface and infect the plant tissue used as trap plant tissue (Figure 2). Some proportions used to mix substrate and water are 1:2, 2:3.5 (weight: volume). This method has several advantages over tissue insulation, since large amounts of soil can be analyzed, increasing the chances of detecting the pathogen when the inoculum concentration is low.

Figure 2 Rhododendron leaves used as plant tissue trap to detect the presence of Phytophthora in substrate or soil samples, in Petri dishes.
It is also more feasible to detect oospores in homotactic species (Erwin and Ribeiro, 1996; Balci et al., 2007; Fichtner et al., 2007). Zoospore-colonized trap tissues are subsequently washed with tap water or sterile distilled water to remove bacteria from the tissue surface, dried with sterile paper and seeded in selective medium (Ferguson and Jeffers, 1999; Jung et al., 2000; Balci et al., 2007; Fitchtner et al., 2007; Hwang et al., 2008; Ghimire et al., 2009). The petiole of Rhododendron leaves is the tissue that is generally infected first, so that it can be cut and planted in selective medium (Figure 3).

Figure 3 Necrotized petioles sectioned from Rhododendron leaves, previously placed with soil or planting substrate and transferred to selective médium NARPH prepared with V-8 agar.
The collection of samples for purposes of detection or population density should be performed in the different seasons of the year since populations may vary from one station to another (Erwin and Ribeiro, 1996). Most species of Phytophthora are favored by conditions of high humidity, temperatures between 15 and 35 °C, humid climates with abundant rains and nutrient-poor substrate or soil. When these conditions are present, it is advisable to analyze samples of the substrate or soil to detect the pathogen (Erwin and Ribeiro, 1996; Drenth and Sendall, 2001; Kong et al., 2009).
It is suggested that the collections are made from moist soil, near apparently healthy roots and from those that are growing (Erwin and Ribeiro, 1996; Balci et al., 2007; Fichtner et al., 2007). Samples should not be exposed to temperatures above 45 °C or to freezing temperatures to maintain the viability of the pathogen (Drenth and Sendall, 2001). They should be placed in plastic bags to prevent dehydration and deposit in a cooler for transportation to the laboratory; placing an insulation material to avoid direct contact of the samples with the ice. It is recommended to keep them moist, at a temperature between 10 and 20 °C and should be processed as soon as possible (Drenth and Sendall, 2001).
Vegetable trap tissues
For the detection of Phytophthora to be more effective from soil and water it is recommended the use of vegetal tissues trap. Selection of tissue type is important for the recovery of Phytophthora species. The trap vegetable tissues that are used can be of different plants and different plant parts such as: cotyledons, small leaf discs, fruits, complete leaves, petals, seedlings, seeds and seed pods. The choice of plant tissue trap depends on the species of Phytophthora suspected to be the causal agent of the disease. The leaves of Rhododendron, Camellia, Quercus, Lupinus, Juniperus needles, apple, pear and cucumber fruits are the most used by which a large number of Phytophthora species can be detected (Erwin and Ribeiro, 1996; Ferguson and Jeffers, 1999; Ivors et al., 2004; Balci et al., 2007; Gevens et al., 2007; Sutton et al., 2009; Reeser et al., 2011; Tooley and Carras, 2011; Themann et al., 2002; Huai et al., 2013). A greater number of Phytophthora species are recovered when whole leaves of the plant tissue trap are used (Ferguson and Jeffers, 1999; Ghimire et al., 2009), than when discs of leaves are used, since it is not recommended because they are easily colonized for Pythium and bacteria.
Purification of bacteria contaminated isolates
To make storage of the strains and studies of morphological and genetic characterization of Phytophthora it is a requirement to obtain isolates free of bacterial contamination. Isolates that are obtained in media with antibiotics are sometimes not free of bacteria. It is recommended to add more than one antibiotic to the medium used to make the isolates (Erwin and Ribeiro, 1996; Nesbitt et al., 1981) to inhibit the growth of different types of bacteria. On the other hand the use of antibiotics can induce resistance in the bacteria and its use in high concentrations inhibits the germination of Phytophthora spores (Erwin and Ribeiro, 1996).
Some alternative methods to the use of antibiotics and low cost are the method of overlapping and the use of acidified medium (Solache-Huacuz et al., 2010; Martin et al., 2012). The first consists of placing a cube of medium with contaminated mycelium, with mycelial growth upwards, in an empty sterile Petri dish, on the cube a triangle of sterile medium is placed (Figure 4).
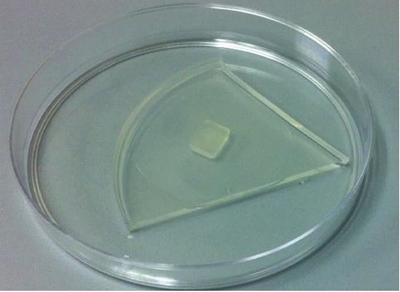
Figure 4 Purification of a Phytophthora isolate contaminated with bacteria, a piece of contaminated mycelium can be placed under a triangle of sterile medium.
The mycelium will grow through the medium triangle towards the bacteria-free surface. The second consists of acidifying the culture medium to pH 3, using 0.14% tartaric acid (Koike, 2007; Solache-Huacuz et al., 2010) or PDA (potato dextrose agar) at pH 4 with 25% lactic acid (Erwin y Ribeiro, 1996). When the mycelium begins to grow, a test must be performed to confirm its purity in Luria Bertani liquid medium (LB) in test tubes, or if there is some other medium that favors the growth of bacteria can also be used. Medium cubes are cut with mycelium, placed inside tubes and incubated at room temperature for 24-48 h (Erwin and Ribeiro, 1996; Solache-Huacuz et al., 2010). Before storing the isolates or performing some study it is necessary to carry out this test, to ensure that they are pure (Figure 5).
Morphological identification
Species identification is performed with the support of tabular cues (Waterhouse et al., 1963; Martin et al., 2012). For this, the cultures to be identified must be pure, that is to say obtained by zoospores or hyphal tip. The genus Phytophthora, is considered difficult to identify especially for those who have no experience (Waterhouse et al., 1963), since the morphological differences between some species of Phytophthora are small and the morphological characters are influenced by the environment, and in some species such as P. capsici present sporangia with different forms. Also, the increasing number of species described, that some species of Phytophthora do not produce sporangia in the media that are commonly used, and the natural presence of hybrids between species of Phytophthora (Man in ´t Veld et al., 2007), complicates the morphological identification.
The production of sporangia can be induced in several ways: a) by adding sterile distilled water to disks or blocks of V-8 medium with mycelium; b) adding an unsterilized soil extract suspension; and c) adding saline to the blocks (Eye et al., 1978; Schoulties et al., 1980; Fichtner et al., 2012; Almaraz-Sanchez et al., 2013).The choice of liquid to be added will depend on the species of Phytophthora being studied. For example, P. cinnamomi suggests using non-sterile soil extract, incubating the isolates under fluorescent white light and performing fluid changes until sporangia are observed (Chen and Zentmyer, 1970; Eye et al., 1978; Ahumada et al., 2013).
Rainwater can also be used to induce sporulation. To perform species identification, there is currently a tabular key published by Martin et al. (2012) and a lucida key available on the internet that includes most morphological features and species (http://idtools.org:8080/key-server/player.jsp?keyId=48). For identification of species, these keys take into account characteristics of asexual spores: sporangium type and shape, dimensions (length and width), number of papillae, expiration, pedicel length, base shape and some sporangiophore characteristics as type of branching, as for sexual spores, type of anteridium, characteristics and dimensions of oogonium and oospora, some additional characteristics for the identification of species of the genus Phytophthora are the presence or absence of chlamydospores, temperature range at which they grow optimal growth temperature, presence or not of swelling in the mycelium and type of reproduction of the strain (homotallic or heterothal) (Martin et al., 2012).
Molecular identification
Modern molecular techniques based on the sequencing of specific regions of DNA are useful tools for the identification of Phytophthora species, since they are an indispensable complement of morphological characterization. These techniques are necessary because of how complicated it can be to determine the morphological characteristics of Phytophthora isolates, for the reasons mentioned above. However, some laboratories do not have the resources for equipment and materials needed to carry out this type of study, so it remains a limitation in many developing countries.
The locus with the highest number of sequences in the databases including all the Phytophthora species described, is the region of the internal transcribed spacers (ITS). The oligonucleotides used primarily for the amplification of the ITS region are ITS4 and ITS6 (White et al., 1990; Cooke et al., 2000; Martin et al., 2012 ). However, the sequences obtained from this locus do not always allow detecting interspecific variation in species closely related phylogenetically, so it is advisable for a correct identification to sequence several nuclear loci. Other commonly used loci are nuclear β-tubulin, and mitochondrial cytochrome c oxidase (CO1, cox2) (Bilodeau et al., 2007; Blair et al., 2008; Robideau et al., 2011). In addition, the real-time PCR technique has provided results that allow the detection and quantification of the pathogen directly in the host (Lees et al., 2012, Engelbrecht et al., 2013).
The development of new technologies in mass sequencing of nucleic acids as metagenomics, allow the detection and discovery of plant pathogens. The new and third generation sequences are relatively new technologies of massive nucleic acid sequencing very useful for the diagnosis and identification of Phytophthora species. Which create profiles of large amounts of sequences allowing to reveal each and every one of the organisms present in the sample (Espindola et al., 2015). These technologies are becoming less expensive, which will allow species identification at the genome level in the near future.
The databases that are considered reliable for identification of Phytophthora isolates through sequences can be found at the following links: http://www.phytophthoradb.org, http://www.phytophthora-id.org and http://www.q-bank.eu, since they have confirmed sequences with which they can be compared by BLAST analysis (Martin et al., 2012).
Conclusions
Morphological and molecular identification is complemented, particularly when new species are found. Using only molecular techniques allows finding a greater number of species; however, it is not possible to characterize them morphologically, which is a limitation for their description. Also, timely diagnosis and correct identification of Phytophthora species are essential to prevent the spread of the pathogen, which will contribute to the proper management of the diseases they cause.
Literatura citada
Ahumada, R.; Rotella, A.; Slippers, B. and Wingfield, M. J. 2013. Pathogenicity and sporulation of Phytophthora pinifolia on Pinus radiata in Chile. Australas. Plant Pathol. 42(4):413-420. [ Links ]
Almaraz-Sánchez, A.; Alvarado-Rosales, D. and Saavedra-Romero, LdeL. 2013. Trampeo de Phytophthora cinnamomi en bosque de encino con dos especies ornamentales e inducción de esporulación. Rev. Chapingo Ser. Cien. Fores. Amb. 19(1):5-12. [ Links ]
Balci, Y.; Balci, S.; Eggers, J.; MacDonald, W. L.; Juzwick, J.; Long, R. P. and Gottschalk, K. W. 2007. Phytophthora spp. associated with forest soils in eastern and north-central U. S. oak ecosystems. Plant Dis. 91(6):705-710. [ Links ]
Bilodeau, G. J.; Lévesque, C. A.; de Cock, A. W. A. M.; Duchaine, C.; Brière, S.; Uribe, P.; Martin, F. N. and Hamelin, R. C. 2007. Molecular detection of Phytophthora ramorum by real-time polymerase chain reaction using taqMan, SYBR green and molecular beacons. Phytopathology. 97(5):632-642. [ Links ]
Blair, J. E.; Coffey, M. D.; Park, S. Y.; Geiser, D. M. and Kang, S. 2008. A Multi-locus phylogeny for Phytophthora utilizing markers derived from complete genome sequences. Fungal Genet Biol. 45(3):266-277. [ Links ]
Borines, L. M.; Palermo, V. G.; Guadalquiver, G. A.; Dwyer, C.; Drenth, A.; Daniel, R. and Guest, D. I. 2014. Jackfruit decline caused by Phytophthora palmivora (Butler). Australas. Plant Path. 43(2):123-129. [ Links ]
Brasier, C. M. 2008. The biosecurity threat to the UK and global environment from international trade in plants. Plant Pathol. 57(5):792-808. [ Links ]
Carter, R. 2004. Arresting Phytophthora dieback. The biological bulldozer. A report by the World Wildlife Fund, (Gland, Switzerland) and the WA Dieback Consultative Council, Western Australia. http://awsassets.wwf.org.au/downloads/sp082_arresting_phytophthora_dieback_1sep04.pdf. [ Links ]
Chen, D. W. and Zentmyer, G. A. 1970. Production of sporangia by Phytophthora cinnamomi in axenic culture. Mycol. 62(2):397-402. [ Links ]
Cooke, D. E. L.; Drenth, A.; Duncan, J. M; Wagels, G. and Brasier, C. M. 2000. A molecular phylogeny of Phytophthora and related oomycetes. Fungal Genet. Biol. 30(1):17-32. [ Links ]
Copes, W. E.; Yang, X. and Hong, C. X. 2015. Phytophthora species recovered from irrigation reservoirs in Mississippi and Alabama nurseries and pathogenicity of three new species. Plant Dis. 99(10):1390-1395. [ Links ]
Dart, N. L. and Chastagner, G. A. 2007. Estimated economic losses associated with the destruction of plants owing to Phytophthora ramorum quarantine efforts in Washington state. Plant Health Progress. [ Links ]
Dart, N. L. and Chastagner, G. A. 2007. High recovery rate of Phytophthora from containerized nursery stock pots at a retail nursery highlights potential for spreading exotic oomycetes. Plant Health Progress. 3-4 pp. [ Links ]
Dart, N. L.; Chastagner, G. A.; Rugarber, E. F. and Riley, K. L. 2007. Recovery frequency of Phytophthora ramorum and other Phytophthora spp. in the soil profile of ornamental retails nurseries. Plant Dis. 91(11):1419-1422. [ Links ]
Davison, E. M. and Tay, F. C. S. 2005. How many soil samples are needed to show that Phytophthora is absent from sites in the south-west of western Australia? Australas. Plant Path. 34(3):293-297. [ Links ]
Drenth, A. and Sendall, B. 2001. Practical guide to detection and identification of Phytophthora. Tropical Plant Protection 1-41. https://research.cip.cgiar.org/confluence/download/attachments/37192003/drenth_phytophthora_practical_guide9.pdf?version=1&modificationdate=1273703622000. [ Links ]
Drenth, E. A. and Guest, D. I. 2004. Diversity and management of Phytophthora in southeast Asia. ACIAR Monograph No. 114, 238p. http://aciar.gov.au/files/node/598/mn114_part1.pdf. [ Links ]
Erwin, D. C. and Ribeiro, O. K. 1996. Phytophthora diseases worldwide. Am. Phytopathol. Soc. St. Paul, MN.USA. 562 p. [ Links ]
Espindola, A.; Schneider, W.; Hoyt, P. R.; Marek, S. M. and Garzon, C. 2015. A new approach for detecting fungal and oomycete plant pathogens in next generation sequencing metagenome data utilizing electronic probes. Int. J. Data Min. Bioinform. 12(2):115-128. [ Links ]
Eye, L. L.; Sneh, B. and Lockwood, J. L. 1978. Factors affecting zoospore production by Phytophthora megasperma var. sojae. Phytopathology. 68:1766-1768. http://www.apsnet.org/publications/phytopathology/backissues/documents/1978articles/phyto68n12-1766.pdf. [ Links ]
Fang, Y. and Ramasamy, R. P. 2015. Current and prospective methods for plant disease detection. Biosensors. 5(3):537-561. [ Links ]
Ferguson, A. J. and Jeffers, S. N. 1999. Detecting multiple species of Phytophthora in container mixes from ornamental crop nurseries. Plant Dis. 83(12):1129-1136. [ Links ]
Fichtner, E. J.; Lynch, S. C. and Rizzo, D. M. 2007. Detection, distribution, sporulation, and survival of Phytophthora ramorum in a California Redwood-Tanoak forest soil. Phytopathol. 93(10):1366-1375. [ Links ]
Fichtner, E. J.; Rizzo, D. M.; Kirk, S. A. and Webber, J. F. 2012. Infectivity and sporulation potential of Phytophthora kernoviae to select North American native plants. Plant Pathol. 61(2):224-233. [ Links ]
Gevens, A. J.; Donahoo, R. S.; Lamour, K. H. and Hausbeck, M. K. 2007. Characterization of Phytophthora capsici from Michigan Surface Irrigation Water. Phytopathol. 97(4):421-428. [ Links ]
Ghimire, S. R.; Richardson, P. A.; Moorman, G. W.; Lea-Cox, J. D.; Ross, D. S. and Hong, C. X. 2009. An in-situ baiting bioassay for detecting Phytophthora species in irrigation runoff containment basins. Plant Pathol. 58(3):577-583. [ Links ]
Hall, K. M. and Albers, H. J. 2009. Economic analysis for the Impact of Phytophthora ramorum on Oregon forest industries. http://www.suddenoakdeath.org/?bibliography=economic-analysis-for-the-impact-of-phytophthora-ramorum-on-oregon-forest-industries. [ Links ]
Hansen, E. M.; Reeser, P. W. and Sutton, W. 2012. Phytophthora beyond agriculture. Annu. Rev. Phytopathol. 50(1):359-378. [ Links ]
Hardham, A. R. 2005. Phytophthora cinnamomi. Mol. Plant Pathol. 6(6):589-604. [ Links ]
Hong, C. X.; Richardson, P. A.; Kong, P. and Bush, E. A. 2003. Efficacy of chlorine on multiple species of Phytophthora in recycled nursery irrigation water. Plant Dis. 87(10):1183-1189. [ Links ]
Hong, C.; Richardson, P. A. and Kong, P. 2002. Comparison of membrane filters as a tool for isolating pythiaceous species from irrigation water. Phytopathol. 92(6):610-616. [ Links ]
Hong, C.; Richarson, P. A. and Kong, P. 2008. Pathogenicity to ornamental plants of some existing species and new taxa of Phytophthora from irrigation water. Plant Dis. 92(8):1201-1207. [ Links ]
Huai, W. X.; Tian, G.; Hansen, E. M. Zhao, W. X.; Goheen, E. M.; Grünwald, N. J. and Cheng, C. 2013. Identification of Phytophthora species baited and isolated from forest soil sand streams in northwestern Yunnan province, China. For. Path. 43(2):87-103. [ Links ]
Hwang, J. Oak, S. W. and Jeffers, S. N. 2008. Detecting Phytophthora ramorum and other species of Phytophthora in streams in natural ecosystems using baiting and filtration methods. Proceedings of the Sudden Oak Death Third Science Symposium. Santa Rosa, CA. http://www.fs.fed.us/psw/publications/documents/psw_gtr214/psw_gtr214_055-058_hwang.pdf. [ Links ]
Ivors, K. L. 2016. Laboratory protocols for Phytophthora species. American Phytopathological Society. http://apsjournals.apsnet.org/action/showBook?doi=10.1094%2F9780890544969). [ Links ]
Ivors, K. L.; Hayden, K. J.; Bonants, P. J. M.; Rizzo, D. M. and Garbelotto, M. 2004. AFLP and phylogenetic analyses of North American and European populations of Phytophthora ramorum. Mycol. Res. 108(4):378-392. [ Links ]
Jeffers, S. N. and Martin, S. B. 1986. Comparison of two media selective for Phytophthora and Pythium species. Plant Dis. 70:1038-1043. [ Links ]
Jeffers, S. N. and Aldwinckle, H. S. 1987. Enhancing Detection of Phytophthora cactorum in naturally infested soil. Phytopathol. 77:1475-1482. [ Links ]
Jiang, R. H. Y. and Tyler, B. M. 2012. Mechanisms and evolution of virulence in oomycetes. Annu. Rev. Phytopathol. 50(1): 295-318. [ Links ]
Jung, T. and Blaschke, M. 2004. Phytophthora root and collar rot of alders in Bavaria: distribution, modes of spread and possible management strategies. Plant Pathol. 53(2): 197-208. [ Links ]
Jung, T. Blaschke, H. and Oßwald, W. 2000. Involvement of soilborne Phytophthora species in Central European oak decline and the effect of site factors on the disease. Plant Pathol. 49(6):706-718. [ Links ]
Jung, T. and Burgess, T. I. 2009. Re-evaluation of Phytophthora citricola isolates from multiple woody hosts in Europe and North America reveals a new species, Phytophthora plurivora sp. nov. Persoonia 22:95-110. [ Links ]
Kasuga, T.; Kozanitas, M.; Bui, M.; Hüberli, D.; Rizzo, D. M. and Garbelotto, M. 2012. Phenotypic diversification is associated with host-induced transposon derepression in the sudden oak death pathogen Phytophthora ramorum. PLoS One. 7(4):34728. [ Links ]
Koike, S. T. Gladders, P. and Paulus, A. O. 2007. Vegetable diseases. A color handbook. Manson Publishing, Ltd. London. 448 p. [ Links ]
Kong, P.; Moorman, G. W.; Lea-cox, J. D.; Ross, D. S.; Richarson, P. A. and Hong, C. 2009. Zoosporic tolerance to pH stress and its implications for Phytophthora species in aquatic ecosystems. Appl. Environ. Microbiol. 75(13):4307-4314. [ Links ]
Kroon, L. P. N. M.; Brouwer, H.; de Cock, A. W. A. M. and Govers, F. 2012. The genus Phytophthora anno 2012. Phytopathol. 102(4):348-364. [ Links ]
Lamour, K.H.; Stam, R.; Jupe, J. and Huitema, E. 2012. The oomycete broad-host-range pathogen Phytophthora capsici. Mol. Plant Pathol. 13(4):329-337. [ Links ]
Loyd, A. L.; Benson, D. M. and Ivors, K. L. 2014. Phytophthora populations in nursery irrigation water in relationship to pathogenicity and infection frecuency of Rhododendron and Pieris. Plant Dis. 98(9):1213-1220. [ Links ]
MacDonald, J. D.; Ali-Shtayeh, M. S.; Kabashima, J. and Stites, J. 1994. Ocurrence of Phytophthora species in recirculated nursery irrigation effluents. Plant Dis.78:607-611. [ Links ]
Man in‘t Veld, W. A.; de Cock, A. W. A. M. and Summerbell, R. C. 2007. Natural hybrids of resident and introduced Phytophthora species proliferating on multiple new hosts. Eur. J. Plant Pathol. 117(1):25-33. [ Links ]
Martin, F. N.; Abad, Z. G.; Balci, Y. and Ivors, K. 2012. Identification and detection of Phytophthora: reviewing our progress, identifying our needs. Plant Dis. 96(8):1080-1103. [ Links ]
Miller, S. A.; Bhat, R. G. and Schmitthenner, A. F. 1994. Detection of Phytophthora capsici in pepper and cucurbit crops in Ohio with two commercial inmmunoassay kits. Plant Dis. 78:1042-1046. [ Links ]
Moralejo, E.; Pérez-Sierra, A. M.; Álvarez, L. A.; Belbahri, L.; Lefort, F. and Descals, E. 2009. Multiple alien Phytophthora taxa discovered on diseased ornamental plants in Spain. Plant Pathol. 58(1):100-110. [ Links ]
Nesbitt, H. J.; Malajczuk, N. and Glenn, A. R. 1981. Bacterial colonization and lysis of Phytophthora cinnamomi. Transactions of the British Mycological Society. 77(1):47-54. [ Links ]
Robideau, G. P.; De Cock, A. W. A. M.; Coffey, M. D.; Voglmayr, H.; Brouwer, H.; Bala, K.; Chitty, D. W.; Désaulniers, N.; Eggertson, Q. A.; Gachon, C. M.; Hu, C. H.; Küpper, F. C.; Rintoul, T. L.; Sarhan. E.; Verstappen, E. C.; Zhang, Y.; Bonants, P. J.; Ristaino, J. B. and Lévesque, C. A. 2011. DNA barcoding of oomycetes with cytochrome c oxidase subunit I and internal transcribed spacer. Mol. Ecol. Resour. 11(6):1002-1011. [ Links ]
Roy, S. G. and Grünwald, N. J. 2014. The plant destroyer genus Phytophthora in the 21st century. Rev. Plant Pathol. 6:388-412. [ Links ]
Schoulties, C. L.; Baker, K. F. and Sabersky-Lehmann, C. 1980. Factors influencing zoospore production by Phytophthora cinnamomi in axenic cultui. Can. J. Bot. 58(19):2117-2122. [ Links ]
Shearer, B. L.; Crane, C. E. and Cochrane, A. 2004. Quantification of the susceptibility of the native flora of the South-West Botanical Province, Western Australia, to Phytophthora cinnamomi. Aust. J. Bot. 52(4):435-443. [ Links ]
Solache-Huacuz, E.; Rodríguez-Alvarado, G.; Naranjo-Bravo, A. E.; Díaz-Celaya, M. y Fernández-Pavía, S. P. 2010. Técnicas de purificación de aislamientos de Phytophthora contaminados por bacterias. Biológicas. 12(1):61-64. [ Links ]
Stewart-Wade, S. M. 2011. Plant pathogens in recycled irrigation water in commercial plant nurseries and greenhouses: Their detection and management. Irrigation Sci. 29(4):267-297. [ Links ]
Streito, J. C.; Jarnouen de Villartay, G. and Tabary, F. 2002. Methods for isolating the alder Phytophthora. Forest Pathol. 32(3):193-196. [ Links ]
Sutton, W.; Hansen, E. M.; Reeser, P. and Kanaskie, A. 2007. Comparing Phytophthora ramorum diagnostic protocols for the national sudden oak death stream monitoring program. Proceedings of the Sudden Oak Death Third Science Symposium. Santa Rosa, California. http://www.fs.fed.us/psw/publications/documents/psw_gtr214/psw_gtr214_461-466_sutton.pdf?. [ Links ]
Sutton, W.; Hansen, E. M.; Reeser, P. W. and Kanaskie, A. 2009. Stream monitoring for detection of Phytophthora ramorum in Oregon tanoak forests. Plant Dis. 93(11):1182-1186. [ Links ]
Themann, K.; Werres, S.; Lüttmann, R. and Diener, H. A. 2002. Observations of Phytophthora spp. in water recirculation systems in commercial hardy ornamental nursery stock. Eur. J. Plant Pathol. 108(4):337-343. [ Links ]
Tooley, P. W. and Carras, M. M. 2011. Enhanced recovery of Phytophthora ramorum from soil following 30 days of storage at 4°C. J. Phytopathol. 159(9):641-643. [ Links ]
Tsao, P. H. and Ocana, G. 1969. Selective isolation of species of Phytophthora from natural soils on an improved antibiotic medium. Nature. 223:636-638. [ Links ]
Tsopmbeng, G. R.; Fontem, D. A. and Yamde, K. F. 2012. Evaluation of culture media for growth and sporulation of Phytophthora colocasiae Racib., causal agent of taro leaf blight. Int. J. Biol. Chem. Sci. 6(4):1566-1573. [ Links ]
Waterhouse, G. M. 1963. Key to the species of Phytophthora de Bary. Commonw. Mycol Inst. Kew, UK. 92 p. [ Links ]
White, T. J.; Bruns, T.; Lee, S. and Taylor, J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis, M. A.; Gelfand, D. H.; Shinsky, J. J. and White, T. J. (Eds.). PCR protocols: a guide to methods and applications. Academic Press, Inc. USA. 482 p. [ Links ]
Zappia, R. E.; Hüberli, D.; Hardy, G. E. St. J. and Bayliss, K. L. 2014 Fungi and oomycetes in open irrigation systems: knowledge gaps and biosecurity implications. Plant Pathol. 63(5):961-972. [ Links ]
Received: August 2017; Accepted: October 2017











 texto en
texto en