Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Revista mexicana de ciencias agrícolas
versión impresa ISSN 2007-0934
Rev. Mex. Cienc. Agríc vol.8 no.6 Texcoco ago./sep. 2017
Articles
Identifying hybrids of Citrus aurantifolia×Citrus limon using simple sequence repeats (SSR) markers
1INIFAP-Campo Experimental Tecomán. Carretera Colima-Manzanillo km 35, Tecomán, Colima, México. (garcia.karina@inifap.gob.mx; orozco.mario@inifap.gob.mx).
2INIFAP-Centro Nacional de Recursos Genéticos. (guzman.luis@inifap.gob.mx).
3Centro Universitario de Ciencias Biológicas y Agropecuarias. (pandreapalmeros@gmail.com).
Apomixis is a type of asexual reproduction where the seeds formation carries genetically identical embryos to the parent, constituting an obstacle in breeding programs for many plant species, including citrus. The identification of hybrid plants is carried out by morphological characters, isoenzymatic assays and molecular markers. The latter have been used more frequently because of their accuracy, standind out the use of random amplified polymorphic DNA (RAPD) and Simple Sequence Repeats (SSR). In Mexican lime (C. aurantifolia) RAPD markers have only been used for hybrid identification, so there are no reports that use SSR markers for this purpose. The aim of this paper was to identify hybrids derived from controlled pollination between C. aurantifolia var. “Colimex”×C. limon var. “Rosenberg” and its reciprocal using SSR molecular markers. During the 2014-2016 years, leaves of lime trees of approximately 12 months of age were collected, which are established in the Campo Experimental Tecomán of the INIFAP. A total of eight SSR molecular markers were evaluated on the progenitors used in this study and the TAA45 and cAGG09 oligonucleotides were selected for the hybrids identification in the two progeny populations. From a total of 40 and 43 F1 individuals from the bi-directional cross between “Colimex”דRosenberg”, 17 and 35 hybrid plants were identified, respectively. The results indicate that SSR markers are efficient and reliable for the identification of Mexican lime hybrids.
Keywords: apomixis; citrus; microsatellites; molecular markers
La apomixis es un tipo de reproducción asexual donde la formación de semillas porta embriones genéticamente idénticos al progenitor, constituyendo un obstáculo en programas de mejoramiento genético de muchas especies vegetales, incluyendo cítricos. La identificación de plantas híbridas se realiza mediante caracteres morfológicos, ensayos isoenzimáticos y marcadores moleculares. Estos últimos se han utilizado con mayor frecuencia debido a su precisión, destacando el uso del DNA polimórfico amplificado al azar (RAPD, “Random Amplified Polymorphic DNA”) y Secuencias Simples Repetidas (SSR, Simple Sequence Repeats). En limón mexicano (C. aurantifolia) únicamente se han utilizado marcadores RAPD para la identificación de híbridos, por lo que no existen reportes que hagan uso de marcadores SSR para este fin. El objetivo del presente trabajo fue identificar híbridos derivados de la polinización controlada entre C. aurantifolia var. “Colimex” ×C. limon var. “Rosenberg” y su recíproca utilizando marcadores moleculares SSR. Durante el año 2014- 2016 se colectaron hojas de árboles de limón de aproximadamente 12 meses de edad, que se encuentran establecidos en el Campo Experimental Tecomán del INIFAP. Se evaluaron en total ocho marcadores moleculares SSR sobre los progenitores utilizados en este estudio y fueron seleccionados los oligonucleótidos TAA45 y cAGG09 para la identificación de híbridos en las dos poblaciones progenie. De un total de 40 y 43 individuos F1 procedentes de la cruza bidireccional entre “Colimex”דRosenberg”, se lograron identificar 17 y 35 plantas híbridas, respectivamente. Los resultados indican que los marcadores SSR son eficientes y confiables para la identificación de híbridos de limón mexicano.
Palabras clave: apomixis; cítricos; marcadores moleculares; microsatélites
Introduction
Citrus fruits are grown in the tropical and subtropical regions of the world. In México, citrus agro-industry represents one of the most important, generating an economic outflow of more than 375 million dollars (SIAP, 2016). Genetic improvement of this crop is a routine activity carried out in several countries with the aim of improving the quality of the fruit or to achieve tolerance to biotic and/or abiotic stress (Tozlu et al., 1999; Mendoza-Rodríguez et al., 2001; Machado et al., 2011; Omura and Shimada, 2016).
Species of the genus Citrus present polyembryony and apomixis, generating disturbances in the process of sexual reproduction between vegetable organisms, in which multiple embryos are found within a seed or where several embryos derived from the pollen receptor of the parent are originated from nuclear tissue surrounding the zygotic embryo (Koltunow et al., 1996; Kepiro and Roose 2007). Most commercial citrus worldwide propagate as grafted trees with the variety of interest in a rootstock (Wutscher and Hill, 1995; Khan and Kender, 2007).
Most of these rootstocks are apomictic, so if it is required to maintain genetic homogeneity this condition may be advantageous as a process of clonal multiplication. In this way, uniform plants can be produced from seeds at low cost. On the other hand, when it is desirable to generate variability by genetic recombination for obtaining hybrids, with resistance to diseases for example, then apomixis represents an obstacle, since the plants are genetically identical to the pollen receptor parent (Khan and Kender, 2007). For the identification of zygotic plants in citrus, phenotypic markers have been used, which are based mainly on leaf morphology.
Furthermore, isozyme tests and the use of flow cytometry have also been used as tools to perform this type of analysis (Anderson et al., 1991; Ruíz et al., 2000; Viloria et al., 2005); however, these techniques may present some limitations. Nowadays, the use of molecular biology has allowed to expand the tools used for the genetic identification of plant species through the use of molecular markers, among which are the ISSR (Inter Simple Sequence Repeat), RAPD (Random Amplification of Polymorphic DNA) and SSR (Simple Sequence Repeat) (Bastianel et al., 1998; Golein et al., 2011; Yildiz et al., 2013; Mondal and Saha, 2014; Mondal et al., 2015).
Turkey has been reported in the efficient use of AG14 and TAA03 oligonucleotide of SSR markers to identify and eliminate nucellar individuals in hybrid populations resulting from crosses between tangerin varieties (C. reticulata), oranges (Citrus sinensis), grapefruit (C. paradisi) (Yildiz et al., 2013). Recently in India four SSR oligonucleotides with repeats have been used AG: CCSM13, CCSM17, CCSM18 and CCSM147 to identify zygotic and nucellar plants in a population of C. reticulata (Mondal et al., 2015).
In Mexico, the species of C. macrophylla and C. volkameriana are commonly used as rootstocks for propagation and establishment of Mexican lime trees. In 2004, Andrade-Rodríguez et al. (2004) conducted a study to determine the zygotic or nucellar origin in C. volkameriana using RAPD markers, with satisfactory results. Similarly, in the case of Mexican lime (C. aurantifolia), RAPD markers have been successfully used, especially the decamer OPH15 (Mondal and Saha, 2014); without there being reports in which other type of molecular markers, such as SSRs, have been used.
In the INIFAP breeding program, crosses between Mexican limes (C. aurantifolia) and Italian limes (C. limon), are routinely performed since it has been observed that the progeny exhibits some tolerance to HLB bacterial disease (Huanglongbing or yellow dragon disease), which causes one of the main phytosanitary problems affecting productivity in the Pacific region of México. Although there are many reports to identify zygotic citrus plants using molecular markers, in México no studies using SSR markers have been reported to identify hybrid plants of C. aurantifolia. In order to establish changes in the hybrids derived from controlled pollination between C. aurantifolia var. “Colimex”×C. limón var. “Rosenberg” and its reciprocal, the present research was carried out using SSR markers.
Materials and methods
Vegetal material
Healthy-looking leaves of lime trees from two progeny populations were collected to identify individuals of zygotic origin and to confirm the hybrid condition of the plants. C. aurantifolia (♀)×C. limon (♂) cross consisted of 40 individuals, while the C. limon (♀)×C. aurantifolia (♂) cross was constituted by 43 individuals. Sampling was performed on 12 months old trees, which are established in the field within the Mexican lime breeding program of the INIFAP Campo Experimental Tecomán, Colima, México, located at 60 masl.
Extraction of genomic DNA
The method described by Bermúdez-Guzmán et al. (2016) was used, with some modifications. All centrifugations were carried out at 4 °C and 13 500 rpm. The leaves of the sampled plants were washed with running water and soap. Approximately 100-200 mg of foliar tissue were then weighed and pulverized in a mortar containing liquid nitrogen and homogenized with 1 mL of CTAB buffer solution [100 mM of Tris-HCl pH 8, 50 mM of EDTA pH 8, 1.4 M of NaCl, 3% (p/v) of CTAB and 1% (p/v) of PVP]. Subsequently, 10 μL of concentrated 2-β-mercaptoethanol and 5 μL of K proteinase (20 mg mL-1) were added.
Samples were incubated for 45 min at 65 °C in water bath, then mixed by inversion every 5-10 min, then centrifuged for 3 min. The aqueous phase was recovered in a new tube and 5 μL of 10 mg m L-1 RNase A (Sigma Aldrich) was added at 37 °C for 10 min. Subsequently a volume of the phenol:chloroform:isoamyl alcohol solution (25:24:1) was added, and vigorously mixed using a vortex and then centrifuged for 10 min.
The recovered supernatant was placed in 1.5 mL tubes and the DNA precipitated with 0.6 volumes of cold isopropanol and 0.1 volumes of 3 M sodium acetate pH 5.2 at -20 °C for 30 min.The mixture was centrifuged for 10 min, the supernatant was decanted and the DNA was washed with 500 μL of 70% ethanol; the tubes were centrifuged for 2 min the aqueous phase was decanted and the tubes were inverted on absorbent paper to remove excess ethanol. The DNAs were dried at room temperature, resuspended in 80 μL of TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH, 8) and finally stored at -20 °C.
Quantification, purity and integrity of genomic DNA
The DNA concentration (ng μ L-1) was quantified using a spectrophotometer (NanoDrop Thermo Scientific). The purity was measured considering the relations of A260:280 and A260:230. Integrity of nucleic acids was verified by 1% agarose gel electrophoresis (Sigma Aldrich) adding 5 μL of genomic DNA from each sample. The gel was stained with 10 mg m L-1 ethidium bromide (Sigma Aldrich) and visualized with ultraviolet light in a photodocumentator (UVP).
Analysis of SSR markers
Parents C. aurantifolia and C. limon were analyzed with eight sets of oligonucleotides described by Kijas et al. (1997) (Table 1) in order to identify those that generate differential polymorphisms in both species. The selected oligonucleotides were used to analyze the two progeny populations of C. aurantifolia×C. Lemon and its reciprocal.
Amplification reactions by PCR were performed in a volume of 15 μL containing 7.5 μL of RedTaq ReadyMixTM (Sigma Aldrich), 5 μL of genomic DNA (20 ng μ L-1), 1 uL of each oligonucleotide and 0.5 µL molecular grade water. Amplification conditions for PCR were performed in a MultiGene Labnet thermocycler and were described by Kijas et al. (1997) with some modifications: initial denaturation at 94 °C for 5 min followed by 35 cycles of 94 °C for 1 min, 55 °C for 30 s and 72 °C for 1 min. The final extension was 72 °C for 10 min.
Vertical polyacrylamide gel electrophoresis was used under non-denaturing conditions using the “PerfectBlue” double gel system (PeQlab). 1 μL of molecular weight marker of 25 and 50 base pairs (bp) (Bioline) and 3 μL of the PCR product of each sample was loaded. The gels were prepared in a 30 mL volume containing 6% acrylamide/bis-acrylamide (29:1), TBE 1 X buffer, 140 μL of 25% APS, 35 μL of TEMED and sterile distilled water, then run at 200 V for 50 min. Finally stained with silver solution according to Sanguinetti et al. (1994) with some modifications: 10 min in fixing solution (10% ethanol and 0.5% acetic acid), 1 rinse with distilled water, 10 min in staining solution (0.2% AgNO3), one rinse with distilled water, 10 to 15 min in developer solution (3% NaOH and 0.5% formaldehyde) and a final rinse with distilled water. The gels were visualized in a transilluminator with white light for analysis.
Results and discussion
Eight oligonucleotides were evaluated for differential bands in C. aurantifolia and C. limon parents used in this paper (Table 1). The TAA45 and cAGG09 oligonucleotides were selected because they allowed the amplification of polymorphic bands of easy interpretation; four of the oligonucleotides amplified mostly non-informative and low resolution monomorphic bands and therefore were not used for the detection of hybrids in the populations analyzed. TAA41 initiator generated very diffuse bands that were not recorded. On the other hand, the TAA52 primer did not allow the amplification of some segment of DNA in the analyzed samples (Figure 1).

Figure 1 Oligonucleotides assessment of SSR markers from parents DNA. C= Citrus aurantifolia var. “Colimex”.R= Citrus limon var. “Rosenberg”. MPM= 25 and 50 bp molecular weight marker (Bioline).
The oligonucleotides of the TAA series described by Kijas et al. (1997) have been widely used in the detection of hybrids between crossings of several citrus species such as grapefruit (C. paradisi), trifoliate orange (C. trifoliata), sweet orange (Citrus sinensis) and tangerine (C. reticulata) (Ruíz et al., 2000; Ahmad et al., 2012; Yildiz et al., 2013; Mondal et al., 2015). However, there are no reports of its use in Mexican lime (C. aurantifolia) and Italian lemon (C. limon), so these results are the first report on these species to determine its potential as hybrid detectors for genetic improvement programs.
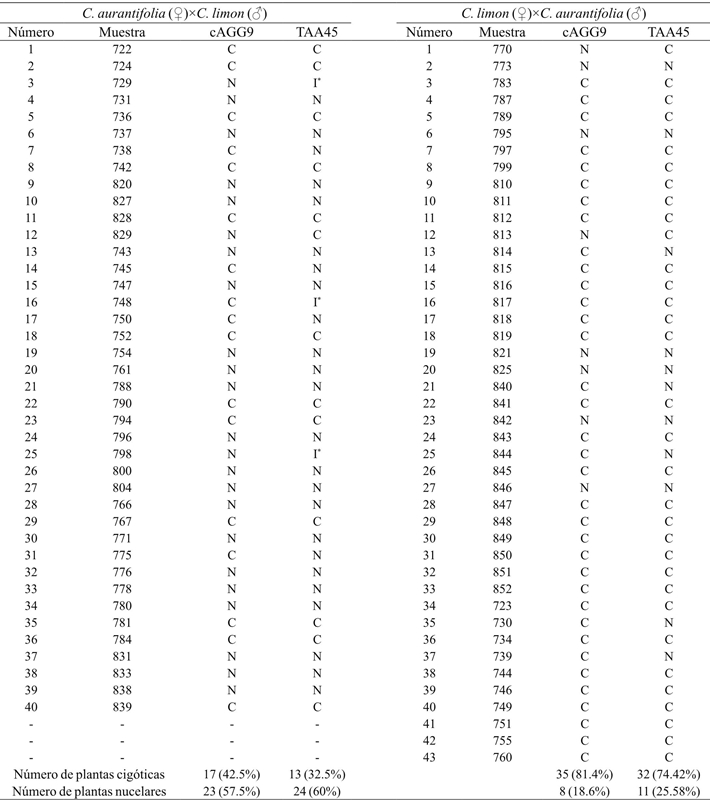
In populations of 40 and 43 individuals from the crosses of C. aurantifolia var. “Colimex”×C. limon var. “Rosenberg” and its reciprocal, respectively, a total of 52 hybrids were detected with the TAA45 and cAGG09 oligonucleotides (Table 2). The TAA45 marker amplified two polymorphic alleles, one present in “Colimex” and absent in “Rosenberg” of approximately 100 bp and another present in “Rosenberg” and absent in “Colimex” of 110 bp. The latter fragment on some gels was visualized as a single band; however, they were three polymorphic bands with very similar molecular weights, reason why sometimes they did not separate. Figure 2a shows the hybrids detected with the TAA45 marker (722, 724, 736, 742 and 828), which also amplified other polymorphic bands that were not taken into account for the identification of hybrids due to their low resolution.
*= no amplificó o presentó barrido el carril del gel.
Table 2 Zygotic or nucellar origin of C. aurantifolia×C. limon populations and its reciprocal by SSR analysis using two pairs of oligonucleotides. C= zygotic; N= nucelar; I= indeterminate.

Figure 2 Electrophoretic profiles obtained with the TAA45 (A) and cAGG09 (B) oligonucleotides. MPM: 25 bp molecular weight marker (Bioline). ♀= pollen receiving plant. ♂= pollen donor plant. C= C. aurantifolia var.“Colimex”. R= C. limon var. “Rosenberg”. N= nucellar origin. C= zygotic origin (hybrids). The numbers correspond to different progeny samples
The cAGG09 primer amplified four polymorphic bands in the analyzed populations, with approximate molecular weights of 90, 100, 110 and 170 bp. In both populations, with this marker, a non-informative 80 bp monomorphic band was amplified. The “Colimex” hybrids were identified by 90 and 170 bp alleles (Figure 2b, lanes 722, 724, 736, 738 and 742), which are present in “Rosenberg” (pollen donor) and absent in “Colimex” (pollen receptor).
Similarly, the “Rosenberg” hybrids were identified by 105 and 115 bp alleles, which are present in “Rosenberg” and absent in “Colimex”. In this sense, it is considered that a hybrid can be determined by identifying a single codominant locus for which the parents do not share alleles, i.e., crosses of the configuration aa×bb, ab×cc or ab×cd.
Also dominant markers, such as RAPD and ISSR, can be used if the pollen receptor is 00 (absent band) and the pollen donor is 11 (homozygous band present) (Kepiro and Roose, 2007). However, confirmation with a second locus is always desirable to confirm the hybrid condition of the individual, which was performed in this research using the TAA45 and cAGG09 markers, whose polymorphisms of each progenitor segregated into the offspring according to the corresponding population.
SSR markers are a useful tool for the study of genetic diversity of citrus by its effectiveness and information generated (Amara et al., 2011; Biswas et al., 2011). Its use for detecting hybrids (Ruíz et al., 2000; Oliveira et al., 2002; Ahmad et al., 2012; Yildiz et al., 2013; Mondal et al., 2015) is more practical, effective and advantageous because the researches that used RAPD and ISSR (Bastianel et al., 1998; Andrade-Rodríguez et al., 2004; Golein et al., 2011) usually get a lot of uninformative monomorphic bands that may hinder the analysis of the data. The RAPD and ISSR, being dominant markers do not allow to differentiate heterozygous individuals unlike the SSR that are codominant markers.
In addition, RAPDs exhibit low reproducibility, as small modifications in the technique such as DNA template concentration, degree of purity and fidelity of the DNA polymerase enzyme, among other factors, may alter the pattern of amplified DNA fragments generated for a sample. Additionally, RAPD are not locus-specific, so electrophoretic band profiles can not be interpreted in terms of loci and alleles and fragments of similar size may not be homologous (Martínez et al., 2010; Kumari and Thakur, 2014).
In this paper bidirectional crossbreeding of C. aurantifolia×C. limon were performed, yielding 42.5% of hybrid plants when “Colimex” was used as pollen receiver, and 81.4% when “Rosenberg” was used for the same purpose (Table 2). Grapefruits, oranges and tangerines have more than 90% of nucellar embryos, depending on the variety. In Mexican lemon, the proportion of nucellar seedlings is 78%, while for Italian lemons it is 32-33% (Frost and Soost, 1968). The results of this research showed slightly lower percentages, 57.5 and 18.6% of nucellar embryos in “Colimex” and “Rosenberg”, respectively. These lower values than previously reported could be due to nutritional factors, as well as genotype-environment interaction (Kepiro and Roose 2007).
In this research, individuals with zygotic origin were considered as hybrids, since the pollinations were controlled, avoiding self-fertilization events. Both markers, cAGG09 and TAA45 coincide in 80 and 83.72% in plants determined as hybrid for C. aurantifolia×C. limon populations and its reciprocal. Samples in which both markers did not coincide with the zygotic or nucellar origin of the progeny were considered the result of cAGG09 oligonucleotide because it amplified well-defined polymorphic bands.
On the other hand, the effectiveness of isoenzymes and SSR markers for detecting hybrids has been compared, concluding that molecular markers are more efficient for this purpose. In this same research they used the oligonucleotides used in this paper: TAA41 and TAA45 obtaining polymorphisms which identified the zygotic origin in populations of C. reticulata×C. sinensis and Poncirus trifoliata self-pollination (Ruíz et al., 2000). Although in this research the use of the TAA41 primer amplified diffuse non-informative bands patterns. In another study, they combined the use of morphological markers assisted by SSR molecular markers of the CCSM series to detect hybrids derived from C. reticulata and C. sinensis (Oliveira et al., 2002).
However, they only confirmed the hybrid status of the plants detected by morphology, so the possible existence of more individuals with a sexual origin that could be detected by the SSR markers is doubted. Meanwhile Ahmad et al. (2012) used the initiators TAA15, TAA27, TAA33 and others from the CCSM series to detect hybrids in three F1 populations of several orange and tangerine crosses, detecting 23 and 5 hybrids of the “NARC 05-18”דTarocco” and NARC 05-17דSanguinello” crosses respectively. In addition, they identified 35 more hybrids of “Kinnow”דTarocco” with the CCSM147 marker. Yildiz et al. (2013) used the primers TAA01, TAA3, TAA41, TAA45, TAA52, CAC33 and cAGG09 to detect hybrids in several populations using tangerines as pollen receptor and crossing them with oranges and grapefruit.
Of the 500 individuals constituting the progenies of the various crosses, primers AG14 and TAA3 were the most effective for identifying zygotic individuals. The “Fremont” and “Robinson” tangerines produced 36.91 and 31.09% of nucellar plants, respectively. Finally, Mondal et al. (2015) recently made use of TAA15, TAA27 and TAA33 oligonucleotides for detecting hybrids of C. reticulata and reported that none of these primers allowed the identification of sexual origin in the evaluated progeny, so they resorted to the initiators of the MRCC series for identification purposes.
Although SSR markers provide useful information for the identification of hybrid plants in different citrus species, for this purpose it is necessary to implement techniques such as diversity array technology (DArT), single nucleotide polymorphism (SNP), or high resolution melt (HRM), however, these are currently very expensive.
Conclusions
SSR markers allowed the efficient identification of zygotic and nucellar plants in progeny populations of Mexican lime “Colimex” and Italian lemon “Rosenberg”. With the use of the cAGG09 primer, the amplification of 4 polymorphic alleles, sufficient to determine the sexual origin of Mexican lemon plants, was identified; however, confirmation with a second locus is always desirable to confirm the hybrid condition of individuals.
A total of 52 hybrid in the populations from crosses C. aurantifolia (♀)×C. limon (♂) and C. limon (♀)×C. aurantifolia (♂) were identified. Additionally, identified hybrids may be infected with HLB in field conditions to determine the level of tolerance to the disease.
Literatura citada
Ahmad, M.; Javaid, A.; Rahman, H.; Hussain, S. I.; Ramzan, A. and Ghafoor, A. 2012. Identification of mandarin×orange hybrids using Simple Sequence Repeat markers. J. Agric. Res.50(2):225-232. [ Links ]
Amara, M. H.; Biswas, M. K.; Zhang, Z. and Guoa, W. W. 2011.Exploitation of SSR, SRAP and CAPS-SNP markers for genetic diversity of citrus germplasm collection. Sci. Hortic. 128(3):220-227. [ Links ]
Anderson, C. M.; Castle, W. S. and Moore, G. A. 1991. Isozymic identification of zygotic seedlings in Swingle citrumelo Citrus paradisi×Poncirus trifoliata nursery and field populations. J.Am. Soc. Hortic. Sci. 116(2):322-326. [ Links ]
Andrade, R. M.; Villegas, M. A.; Gutiérrez, E. M. A.; Carrillo, C. G.and García, V. A. 2004. Polyembryony and RAPD markers for identification of zygotic and nucellar seedlings in Citrus.Agrociencia. 39(4):371-383. [ Links ]
Bastianel, M.; Schwarz, S. F.; Filho, H. D. C.; Lin, L. L.; Machado, M.and Koller, O. C. 1998. Identification of zygotic and nucellar tangerine seedlings (Citrus spp.) using RAPD. Gen. Mol. Biol.21(1):123-127. [ Links ]
Biswas, M. K.; Chai, L.; Amar, M. H.; Zhang, X. and Deng, X. 2011.Comparative analysis of genetic diversity in citrus germoplasm collection using AFLP, SSAP, SAMPL and SSR markers. Sci. Hortic. 129(4):798-803. [ Links ]
Frost, H. B. and Soost, R. K. 1968. Seed reproduction: development of gametes and embryos. In: W. Reuther, L. D. Batchelor and H.J. Webber (Eds.). The citrus industry. University of California Press. Berkeley, California. 1:290-324. [ Links ]
Golein, B.; Fifaei, R. and Ghasemi, M. 2011. Identification of zygotic and nucellar seedlings in citrus interspecific crosses by inter simple sequence repeats (ISSR) markers. Afr. J. Biotechnol.10(82):18965-18970. [ Links ]
Kepiro, J. L. and Roose, M. L. 2007. Nucellar embryony. In: Khan, I.(Eds.). Citrus genetics, breeding and biotechnology. CABI. UK. 141-149 pp. [ Links ]
Khan, I. Q. and Kender, W. J. 2007. Citrus breeding: Introduction and objectives. In: Khan, I. (Eds.). Citrus genetics, breeding and biotechnology. CABI. UK. 1-8 pp. [ Links ]
Kijas, J. M. H.; Fowler, J. C. S.; Thomas, M. R and Roose, M.L.1997. Integration of trinucleotide microsatellites into a linkage map of Citrus. Theoretical and Applied Genetics.94: 701-706. [ Links ]
Koltunow, A. M.; Hidaka, T. and Robinson, S. P. 1996. Polyembryony in citrus. Plant Physiol. 110:599-609. [ Links ]
Kumari, N. and Thakur, S. K. 2014. Random amplified polymorphic DNA. A brief review. Amrican Journal Animal Veter. Sci.9(1):6-13. [ Links ]
Machado, M. A.; Cristofani, Y. M. and Bastianel, M. 2011. Breeding, genetic and genomic of citrus for disease resistance. Rev. Bras.Frutic. Jabot. -SP. 158-172. [ Links ]
Martínez, M. C.; Helguera, M. y Carrera, A. 2010. Marcadores moleculares. In: Levitus, V.; Echenique, G.; Rubinstein, C.;Hopp E. y Mroginski L. (Eds.). Biotecnología y mejoramiento vegetal II. INTA. Argentina. 70-85 pp. [ Links ]
Mendoza, R. M. F.; Cervera, G. M. T.; Cabezas, M. J. A.; Cenis, J. L. y Martínez, Z. J. M. 2001. Utilización de marcadores AFLP y SAMPL en la identificación genética de especies y variedades de cítricos. Biotecnología Vegetal. 1:11-96. [ Links ]
Mondal, B. and Saha, R. 2014. Identification of zygotic and nucellar seedling of Citrus aurantifolia and Citrus reticulata using RAPD. Inter. J. Adv. Biotechnol. Res. 15(1):25-30. [ Links ]
Mondal, B.; Pramanick, S.; Saha, R. and Karmakar, M. 2015. Application of simple sequence repeat markers for demarcation of Citrus reticulata nucellar and hybrid seedlings. Inter. J. Biosci.6(2):128-133. [ Links ]
Oliveira, A. C.; Novac, G. A.; Cristofani, M. and Machado, M. A. 2002.Identification of citrus hybrids through the combination of leaf apex morphology and SSR. Euphytica. 128:337-339. [ Links ]
Omura, M. and Shimada, T. 2016. Citrus breeding, genetics and genomics in Japan. Breed. Sci. 66(1):3-17. [ Links ]
Ruiz, C.; Paz, B. M. and Asíns, M. J. 2000. A quick methodology to identify sexual seedling in citrus breeding programs using SSR markers.Euphytica. 112:89-94. [ Links ]
Sanguinetti, C. J.; Neto, E. D. and Simpson, A. J. G. 1994. Rapid silver staining and recovery of PCR products separated on polyacrylamide gels. Biotechniques. 17(5):915-919. [ Links ]
SIAP. 2016. Atlas agroalimentario 2016. Primera edición. http://www.gob.mx/siap/. [ Links ]
Tozlu, I.; Guy, C. L. and Moore, G. A. 1999. QTL analysis of morphological traits in an intergeneric BC1 progeny of Citrus and Poncirus under saline and non-saline environments. Genome. 42(5):1020-1029. [ Links ]
Viloria, Z. and Grosser, J. W. 2005. Acid citrus fruit improvement via interploid hybridization using allotetraploid somatic hybrid and autotetraploid breeding parents. J. Am. Soc. Hortic. Sci.130(3):392-402. [ Links ]
Wutscher, H. K. and Hill, L. L. 1995. Performance of ‘Hamlin’ orange on 16 rootstocks in east-central Florida. HortScience. 30(1):41-43. [ Links ]
Yildiz, E.; Klaplankiran, M.; Demirkeser, T. H.; Uzun, A. and Toplu, C.2013. Identification of zygotic and nucelar individuals produced from several Citrus crosses using SSRs markers. Notulae Bot.Hor. Agrobot. 41(2):478-484. [ Links ]
Received: March 2017; Accepted: June 2017











 texto en
texto en 



