Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Similares em
SciELO
Similares em
SciELO
Compartilhar
Revista mexicana de ciencias agrícolas
versão impressa ISSN 2007-0934
Rev. Mex. Cienc. Agríc vol.7 no.6 Texcoco Ago./Set. 2016
Articles
Detection of Clavibacter michiganensis ssp. michiganensis by PCR in in tomato plants (Lycopersicon esculentum Mill.)
1Instituto Tecnológico de Roque. Carretera Celaya-Juventino Rosas, km 8, C. P. 38110. Roque municipio de Celaya, Guanajuato. México. (jesus_friasp@yahoo.com).
2Departamento de Ingeniería Bioquímica, Instituto Tecnológico de Celaya, Av. Tecnológico y A. García Cubas S/N, CP 38010, Celaya, Guanajuato. México. Tel: 01 (461) 6117575 Ext. 5475 y Fax. Ext. 5472. (gerardo.acosta@itcelaya.edu.mx; kfersalinas@hotmail.com; lorenzogo@yahoo.com).
3Campo Experimental Bajío- INIFAP. Carretera Celaya-San Miguel de Allende, km 6.5. Celaya, Guanajuato. México. (gonzalez.mario@inifap.gob.mx).
4Facultad de Ingeniería-Universidad Autónoma de Querétaro. C. P 76010. Santiago de Querétaro, Querétaro, México. (ramon.guevara@uaq.mx; torres.irineo@gmail.com).
Bacterial canker caused by Clavibacter michiganensis ssp. michiganensis (Cmm) is a devastating disease of tomato around the world. The aim of this study was to describe a rapid procedure based on PCR for the detection of pathogenic Cmm in tomato plants. The DNA was isolated by boiling method and PCR was performed to amplify pat-1 gene (serine protease) using the oligonucleotides CMM5F and CMM6R whose amplicon size was 608 bp. The fragment was cloned, sequenced and compared with the database from NCBI (National Center for Biotechnology Information), showing 100% identity with sequences pat-1 gene of Cmm. Additionally, from the sequence obtained two pairs of nested oligonucleotides were designed to produce amplicons of 250 bp to an annealing temperature of 62 ° C. The results described in this paper will allow rapid and accurate identification of Cmm by PCR in seed and tomato plant tissue by reducing the analysis time from 24 to 4 h approximately.
Keywords: Clavibacter michiganensis ssp.; michiganensis; Lycopersicon esculentum Mill.; bacterial canker; boiled; nested oligonucleotides
El cáncer bacteriano causado por Clavibacter michiganensis ssp. michiganensis (Cmm), es una enfermedad devastadora del jitomate alrededor del mundo. El objetivo del presente estudio fue describir un procedimiento rápido basado en PCR para la detección de Cmm patogénica en plantas de jitomate. El DNA fue aislado por el método de hervido y se realizó PCR para la amplificación del gen pat-1 (serina proteasa), empleando los oligonucleótidos CMM5F y CMM6R, cuyo tamaño de amplicón fue de 608 pb. El fragmento fue clonado, secuenciado y comparado con la base de datos del NCBI (Centro Nacional para la Información Biotecnológica), mostrando 100% de identidad con secuencias del gen pat-1 de Cmm. Adicionalmente, de la secuencia obtenida se diseñaron 2 pares de oligonucleótidos anidados para producir amplicones de 250 pb a una temperatura de alineamiento de 62 °C. Los resultados descritos en el presente trabajo permitirán la identificación rápida y precisa de Cmm por PCR en semilla y en tejido vegetal de jitomate, al reducir el tiempo de análisis de 24 a 4 h aproximadamente.
Palabras clave: Clavibacter michiganensis ssp.; michiganensis; Lycopersicon esculentum Mill.; cáncer bacteriano; hervido; oligonucleotidos anidados
Introduction
The total production of tomatoes in Mexico is 2.2 million tons in an area of 96 651 ha, is the leading exporter of tomatoes in the world with 1.4 million tons and exports in the order of 2038 million dollars a year, it generates 72,000 direct jobs and 10.7 million indirect jobs (SIAP, 2013; Flores, 2014). Among the factors limiting the production of this crop are phytosanitary problems such as bacterial canker that is caused by Clavibacter michiganensis ssp michiganensis (Cmm), a quarantine disease and with international impact by the economic losses in tomato production, mainly in greenhouses. When present it is devastating and with full dissemination, Cmm can survive in plant debris or in soil for periods of 2 or 3 years, which hinders its control (Fatmi and Schaad, 2002; Flores, 2014).
Cmm is distributed in all areas where tomatoes are grown, there have been epidemics in Israel, Japan, Spain and Mexico on average can cause losses of 80 to 100% (Borboa-Flores et al., 2009; EPPO, 2010; De Leon et al., 2011; NAPPO, 2013). Infected seeds are the primary source of inoculum and are responsible for the occurrence of serious outbreaks of infection. Low levels of contamination of seed are required to start an epidemic, it is difficult to timely diagnose because when infection begins the symptoms take time to appear, and the first signs of bacterial canker usually appear between 30 and 40 days after transplantation, which makes difficult its control. Therefore, it is necessary to have reliable and highly sensitive methods for effective diagnosis (Chang et al., 1992; Milijašević et al., 2007; Milijasevic-Marcic et al., 2012). On the other hand, there have not been developed resistant varieties to this pathogen despite frequent attempts of improvement (Eichenlaub and Gartemann, 2011).
The methods used to detect Cmm are immunofluorescence and ELISA (Riley, 1987; Nemeth et al., 1991; Franken et al., 1993; Kaneshiro and Alvarez, 2001), which determine the presence or absence, but do not diagnose virulence. Other molecular methods of detection include Southern hybridization using probes derived from CelA and pat-1 genes (Dreier et al., 1995) coding for a endocellulase and a serine protease, respectively and from virulence plasmids PCM1 and PCM2 (Dreier et al., 1997); however, such serologic and hybridization procedures have some limitations such as low sensitivity and long waiting times for results.
Pathogen detection in asymptomatic or infected plants when transplanting is difficult, because the symptoms are not observed, this allows its spread and makes hard to control it in seedbed, therefore, it is necessary to optimize a reliable and accurate detection method of Cmm, in plant tissue and even to prevent the spread by the use of infected seed. PCR is sensitive and by designing specific oligonucleotides combined with rapid extraction procedures of DNA (Holmes and Quigley, 1981; Dellaporta et al., 1983; Acosta-Garcia et al., 2012) can determine the identity and suggest the virulence of phytopathogenic bacteria (Dreier et al., 1995; Zhao et al., 2007; Rodríguez, 2013), and avoid false positives and negatives (Dreier et al., 1995; Souza-Santos et al., 1997; Pastrik and Rainey, 1999; Kaneshiro and Alvarez, 2001; Burokiene, 2006).
The objective of this research was to develop a PCR procedure for rapid and accurate identification of Cmm by designing specific nested oligonucleotides and DNA extraction by boiling for amplification of a fragment from virulence pat-1 gene (serine protease) 250 bp.
Materials and methods
Biological material
The isolated strain of Clavibacter michiganensis ssp. michiganensis (Cmm) of Saladette tomato plants from Zamora Michoacán was provided by the phyto-bacterology lab from the Graduate College Campus Montecillo Montecillo, State of Mexico. The Cmm strain was kept on YDC selective medium (yeast extract, dextrose, CaCO3 and agar) and propagated in LB liquid medium (Luria-Bertani) incubated for 48 h at 28 °C. Lycopersicon esculentum Mill plants Saladette type with symptoms and asymptomatic to Cmm, were collected in an open field in the locality "La Rinconada" de Zamora, Michoacán during the spring-summer 2013. For transformation chemically competent cells of Escherichia coli were used (Invitrogen™) TOP 10 F'. Plasmid TOPO TA 4 (Invitrogen™) was used for cloning the PCR product. Restriction enzyme Eco RI was used to release the cloned PCR products.
DNA extraction
To obtain DNA from Cmm the boling method (Holmes and Quigley, 1981) was used. 1.5 mL Eppendorf tubes were prepared, 100 mg of glass beads of 450-600 μm and 150 of TE μL (Tris base, 50 mM EDTA, 2 mM) were added. Then transferred 2-4 mg of bacteria from solid plates with metal handle, in both plant material with symptoms and asymptomatic were transferred from 80-100 mg to each tube, in all cases were agitated with a vortex for 2 min. The tubes were sealed with parafilm, boiled for 5 min at 100 °C and mixed by stirring for 1 min. To each tube were added 150 μL of a phenol: chloroform (1: 1, v: v) mixture, stirred with the aid of a vortex for 1 min and centrifuged at room temperature at 12 000 rpm for 5 min. The supernatant of each sample was transferred to a new sterile Eppendorf tube, adding 100 μL of chloroform, vortexed for 1 min and centrifuged as in the previous step. 1 μL of each supernatant was used as temperate for PCR reaction. As control, DNA extraction was performed by the Dellapota method (Dellaporta et al., 1983) recommended for plants. DNA isolation was verified by electrophoresis in agarose gel at 1.2% stained with GelRed™ (Biotum).
Polymerase chain reaction (PCR)
Amplification of a fragment form pat-1 gene (serine protease) of Cmm was performed by PCR using the oligonucleotides: CMM5F and CMM6R (Dreier et al., 1995; Table 1); and as temperate the DNA obtained by the boiling method (Holmes and Quigley, 1981). The components of the reaction mixture were: 17.4 µL of water, 2.5 µL of reg taq 10 X, 1.5 µL of MgCl2, 1 µL of sense and antisense oligonucleotides, 0.5 µL dNTPs, 0.1 µL Taq and 1.0 µL of temperate DNA for total reaction volume 25 µL . Thermocycler GeneAmp PCR System 2400 Perkin conditions were: Denaturation 94 °C, 3 min, plus 35 cycles with the following temperatures and times: 94 °C, 30 s; alignment 55 °C, 1 min; synthesis 72 °C, 1 min and finally 1 additional cycle at 72 °C, 5 min, to obtain 250 bp amplicons a temperature annealing of 62 °C, for 30 s and synthesis 72 °C, 30 s (Table 1) were used. Amplification of PCR products were verified by electrophoresis on agarose gel 1.2% stained with GelRed™ (Biotum), the molecular size marker GeneRuler 1 kb DNA Ladder (Life Technologies) was used and the image was recorded on a transilluminator Vilber Lourmat (Marne-La-Vallee Cedex, France).
Table 1. Oligonucelotides used in this work.
| Iniciadores | Secuencia | Número | Tm |
|---|---|---|---|
| CMM5F | 5´GCGAATAAGCCCATATCAA3´ | 19 | 55 ºC (Dreier et al., 1995) |
| CMM6R | 5´CGTCAGGAGGTCGCTAATA3´ | 19 | 55 ºC (Dreier et al., 1995) |
| HCMA1S | 5´GCATTGCGGCGGTCGTGGTG3´ | 20 | 62 ºC (Presente estudio) |
| HCMB1As | 5´TCCGGCTACCGGCACGGATG3´ | 20 | 62 ºC (Presente estudio) |
| HCMC2S | 5´CGAAGCATTGCGGCGGTCGTG3´ | 21 | 62 ºC (Presente estudio) |
| HCMD2As | 5´TCCGGCTACCGGCACGGATGA3´ | 21 | 62 ºC (Presente estudio) |
Cloning, sequencing and comparison
The PCR product was isolated and cloned into the vector pCR4-TOPO®TA, colonies for plasmid DNA extraction were selected and digestion with Eco RI enzyme to verify the presence of the insert. Plasmid DNA purification of 2 clones was performed using QIAprep Spin Miniprep Kit Qiagen columns to obtain DNA-in quantity and quality for sequencing. Thus, the A260nm /A280 nm index was determined using the spectrophotometer Thermo ScientificTM NanoDrop 2000/200c, which was close to 2.0. The oligonucleotide sequence was determined using the ABI PRISM 310 automated sequencer (Perkin Elmer, Norwalk, CT, USA). Comparison with the database was performed using the BLASTX algorithm (Altschul et al., 1990) from the National Center for Biotechnology Information (NCBI).
Oligonucleotide Design
For the design of the 2 nested oligonucleotides pairs, melting temperature (Tm) and amplicon size 250 bp from pat-1 gene (serine protease) of Clavibacter michiganensis ssp. michiganensis, the algorithm OligoPerfectTM Designer from Invitrogen (Table 1) was used.
Results
Isolation of DNA Cmm
For identification of the virulent strain of Cmm, DNA extraction by boiling (Holmes and Quigley, 1981) and Dellaporta (Dellaporta et al., 1983) methods were used. The result is shown in Figure 1, where a discrete and upper band in size to10 000 bp corresponding to the genomic DNA of Cmm can be observed. The DNA obtained by the boiling method showed significant contamination by mRNA (lines 1 and 2), which is absent in the isolated DNA samples by the Dellaporta method (lines 3 and 4), on the other hand, the A260nm / A280 nm index for isolated plant DNA by the boiling method was 1.71, while for DNA isolated by Dellaporta method (Dellaporta et al., 1989) was 1.93. Additionally, the DNA isolated by the boiling method showed a concentration of 0.595 μg μL-, as for the obtained by the Dellaporta method was 0.117μg μL- (Table 2), suggesting that samples obtained by both methods can be used as temperate for PCR reactions.

Figure 1. DNA extraction of Clavibacter michiganensis ssp michiganensis (Cmm). Lines: MT, size marker; 1 and 2, DNA obtained by the boiling method; 3 and 4, DNA obtained by the Dellaporta method.
Table 2. Comparative data of 2 methods for genomic DNA extraction for Cmm and tomato plant.
| Método de extracción para Cmm | Tiempo | Material de partida (mg)a | Concentración de DNA (μg μL-)b | Purezad |
| Dellaporta | 19 h | 4 (0.778) | 0.125 (0.051) | 1.91 |
| Hervido | 30 min | 3 (0.408) | 0.576 (0.115) | 1.68 |
| Método de extracción para planta de jitomate | Tiempo | Material de partida (mg) | Concentración de DNA (μg/μL) | Pureza |
| Dellaporta | 19 h | 90 (14.14) | 0.117 (0.058) | 1.93 |
| Hervido | 30 min | 88 (17.79) | 0.595 (0.128) | 1.71 |
Amplification of fragment pat-1 gene of Cmm
To reduce the time of analysis were used for the PCR reaction as temperate DNA of Cmm obtained by the boiling method and published oligonucleotides CMM5F and CMM6R (Dreier et al., 1995; Table 1). The products obtained were separated on agarose gel 1.2% stained with GelRed ™ (Figure 2) showing amplification of a greater band than 500 bp (Line 1), which may correspond to the fragment pat-1 gene (serine protease), which has a size of 614 bp (Dreier et al., 1995). As negative control of PCR reaction (line 2), it was used as the temperate DNA obtained by boiling from healthy tomato plant and CMM5F and CMM6R oligonucleotides.
Identification of Clavibacter michiganensis ssp. michiganensis (Cmm)
To identify the pathogenic strain Cmm, the PCR fragment of approximately 600 bp was cloned into the pCR4-TOPO®TA vector and transformed into Escherichia coli cells chemically competent. Plasmid DNA was purified to obtain the sequence, which is shown in Figure 3 where points out the CMM5F oligonucleotide and complement oligonucleotide CMM6R, suggesting that a fragment of 608 bp from pat-1 gene (serine protease) of Cmm has been cloned. The sequence obtained was compared with the database from the National Center for Biotechnology Information (NCBI) using the BLASTX algorithm (Altschul et al., 1990) that translates the sequence and compares it to the registered proteins, the result is shown in Table 3, where sequences of some proteins of the strains with the respective access numbers are shown: Clavibacter michiganensis ssp. michiganensis (WP_011931204.1), C. michiganensis ssp michiganensis NCPPB 382 (CAM98537.1), C. michiganensis ssp. michiganensis (AEM65933.1), C. michiganensis ssp. michiganensis (AEM65932.1), among others, whose E value was close to 0.0 with pat-1 gene (serine protease) from Clavibacter michiganensis ssp. michiganensis, confirming the identification of pathogenic Cmm.

Figure 3. Base sequence of 608 bp fragment from pat-1 gene (serine protease) of Cmm. The sequences of the oligonucleotides CMM5F and the second chain CMM6R are underlined.
Table 3. Sequences that showed significant alignment (NCBI) with E value close to 0.0.
| Descripción | Valor E | No. de acceso |
|---|---|---|
| Serina proteasa [C. michiganensis ssp michiganensis] | 1e-126 | WP_011931204.1 |
| Serina proteasa [C. michiganensis ssp michiganensis NCPPB 382] | 4e-102 | CAM98537.1 |
| Serina proteasa [C. michiganensis ssp michiganensis] | 4e-75 | AEM65933.1 |
| Serina proteasa [C. michiganensis ssp michiganensis] | 7e-73 | AEM65932.1 |
Nested oligonucleotide design for the production of 250bp amplicons by PCR
Based on the sequence obtained two pairs of nested oligonucleotides to produce 250 bp amplicons were designed with an annealing temperature (Tm of 62 °C) which were designated arbitrarily HCMA1S-HCMB1As (20-mer) and HCMC2S- HCMD2As (21-mer). DNA isolated by the boiling method from tomato plants with symptoms to pathogenic Cmm were employed as temperate for PCR amplification of a fragment of the pat-1 gene (serine protease). The result is shown in Figure 4, where the amplification of a specific band of 250 bp with both pairs of nested oligonucleotides (lines 1 and 2) is observed. As a positive control a PCR was performed using as temperate the DNA of tomato plants with Cmm symptoms and CMM5F and CMM6R oligonucleotides, which generated the expected 608 bp fragment. As negative control, were used as temperate the DNA isolated from asymptomatic tomato plants to Cmm and the pair of nested oligonucleotides HCMA1S-HCMB1As, allowing the development of a rapid and specific PCR procedure, combining DNA extraction by the boiling method (Holmes and Quigley, 1981) and nested oligonucleotides with Tm of 62 °C to produce amplicons of 250 bp from pat-1 gene (serine protease) for the identification of pathogenic Cmm.
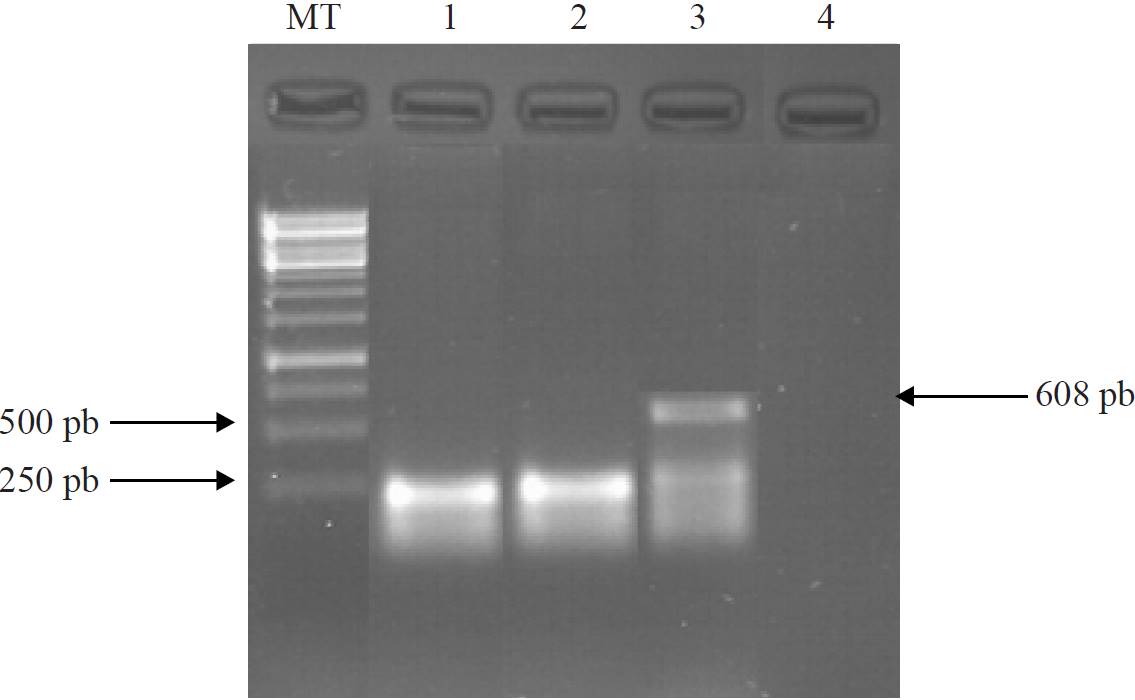
Figure 4. PCR amplification of a 250 bp fragment from pat-1 gene of Cmm from DNA of plants with symptoms. Lines: MT, size marker; 1) nested oligonucleotides HCMA1S and HCMB1As; 2) nested oligonucleotides HCMC2S and HCMD2As; 3) positive control, oligonucleotides CMM5F and CMM6R; and 4) negative control, DNA plant from asymptomatic tomato as temperate and nested oligonucleotides HCMA1S and HCMB1As.
Discussion
In the present study a procedure based on PCR to detect Cmm in tomato plants (Lycopersicon esculentum Mill) Saladette type was developed. To do this it was proposed to combine DNA extraction by boiling method (Holmes and Quigley, 1981) and the use of oligonucleotides HCMA1S-HCMB1As (20-mer) and HCMC2S- HCMD2As (21-mer) with Tm of 62 °C for amplification of 250 bp fragment from pat-1 gene (serine protease) of Cmm. The extraction method by boiling, which consumes a time of about 30 min, the DNA showed a RNA fraction (Figure 1): however, it did not show difference with Dellaporta method (Dellaporta et al., 1983) described for plants, which requires approximately 19 h to completion.
Because in both samples the corresponding band to genomic DNA is present in discrete form of a larger size than 10 000 bp, with an average A260nm / A280 nm index of 1.70 indicating DNA quality (Table 2). The DNA concentration obtained by the boiling method was higher than that obtained by Dellaporta method, the above is justified since the spectrophotometer registers the concentration of nucleic acids, which in this case includes total RNA (Table 2). For PCR amplification of pat-1 gene (Serine protease) of Cmm, by using DNA isolated by boiling as temperate and as primers the CMM5F and CMM6R oligonucleotides (Dreier et al., 1995), amplicon obtained was an upper band to 500 bp (Figure 2, line 1), suggesting that corresponds to the fragment from pat-1 gene (serine protease) which has been reported as a size of 614 bp (Dreier et al., 1995), which was confirmed by sequencing the fragment.
The null amplification from DNA obtained by boiling from asymptomatic tomato plant and CMM5F and CMM6R (Figure 2, line 2) and HCMA1S-HCMB1As (Figure 4, line 4) oligonucleotides confirm the specificity of PCR reaction for Cmm DNA. Reducing the time of analysis to detect Cmm, also is influenced by the use of nested oligonucleotides (HCMA1S-HCMB1As and HCMC2S-HCMD2As) with a Tm of 62 °C and annealing time and synthesis of 30s for production of 250 bp amplicons. These conditions reduce the total time form PCR program, by decreasing the time required for amplification form 4h that were required with the above protocol to 3 h, using as temperate DNA obtained by boiling. In Figure 4 (lines 1 and 2), shows the synthesis of specific fragments of 250bp of pat-1 gene (serine protease).
Conclusions
A fast, simple and accurate method based on PCR to detect pathogenic Cmm that consumes approximately 4 h from the arrival of the sample compared to the conventional method that requires 24 h, which will allow to set in less time, strategies to prevent the spread of bacterial inoculum canker in tomato plants.
Literatura citada
Acosta, G. G.; Pantoja, H. M. A.; Muñoz, S. C. I.; Pérez, P. C.; Guevara, G. R. G.; Torres, P. I.; Delgadillo, S. F.; González, Ch. M. M. y Guevara, O. L. 2012. Transformación del hongo fitopatógeno Sclerotium cepivorum Berk empleando fusión de protoplastos. Rev. Méx. Cienc. Agric. 3(7):1333-1345. [ Links ]
Altschul, S. F.; Gish, W.; Miller, W.; Myers, E. W. and Lipman, D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [ Links ]
Borboa, F. J.; Rueda, P. E.; Acedo, F. E.; Ponce, J. F.; Cruz, M.; Grimaldo, J. O. y García, O. A. M. 2009. Detección de Clavibacter michiganensis subespecie michiganensis en el tomate del estado de Sonora, México. Rev. Fitotec. Mex. 32(4):319-326. [ Links ]
Burokiene, D. 2006. Early detection of Clavibacter michiganensis subespecie michiganensis in tomato seedlings. Agron. Res. 4:151-156. [ Links ]
Chang, R. J.; Ries, S. M. and Pataky, J. K. 1992. Effects of temperature, plant age, inoculum concentration and cultivar on the incubation period and severity of bacterial canker of tomato. Plant Dis. 76:1150-1155. [ Links ]
De León, L.; Siverio, F.; López, M. M. and Rodríguez, A. 2011. Clavibacter michiganensis subsp. Michiganensis a seedborne tomato pathogen: healthy seeds are still the goal. Plant dis. 95(11):1328-1338. [ Links ]
Dellaporta, S. L.; Wood, J. and Hicks, J. B. 1983. A plant DNA minipreparation version II. Plant Mol. Biol. Rep. 18:61-64. [ Links ]
Dreier, J.; Bermpohl, A. and Eichenlaub, R. 1995. Southern hybridization and PCR for specific detection phytopathogenic Clavibacter michiganensis subsp. michiganensis. Phytopathology. 85:462- 468. [ Links ]
Dreier, J.; Meletzus, D. and Eichenlaub, R. 1997. Characterization of the plasmid encoded virulence region pat-1 of phytopathogenic Clavibacter michiganensis subsp. michiganensis. Mol. Plant Microbe Inteact. (102):195-206. [ Links ]
Eichenlaub, R. and Gartemann K. H. 2011. The Clavibacter michiganensis subspecies: molecular investigation of gram-positive bacterial plant pathogens. Department of Genetechnology, Microbiology, Faculty of Biology, University of Bielefeld, Germany. Annu. Rev. Phytopathol. 49:445-464. [ Links ]
EPPO. 2010. Diagnostics. Clavibacter michiganensis subsp. insidiosus. Bulletin 40:353-364. [ Links ]
Fatmi, M. and Schaad N. W. 2002. Survival of Clavibacter michiganensis subsp. michiganensis in infected tomato stems under natural field conditions in California. Ohio, and Morocco. Plant Pathol. 51:149-154. [ Links ]
Flores, D. 2014. Mexico: tomato annual: production down slightly next year while exports up slightly. USDA foreign agricultural service. Gain report number: MX4043. [ Links ]
Franken, A. A. J. M.; Kamminga, G. C.; Snijders, W.; Van Der Zouwen, P. S. and Birnbaun, Y. E. 1993. Detection of Clavibacter michiganensis ssp. michiganensis in tomato seeds by immunofluorescence microscopy and dilution plate. Neth. J. Plant Pathol. 99(3):125-137. [ Links ]
Holmes, D. and Quigley, M. 1981. A rapid boiling method for the preparation of bacterial plasmids. Analyt. Biochem. 114:193-197. [ Links ]
Kaneshiro, W. S. and Álvarez, A. M. 2001. Specificity of PCR and ELISA for hypovirulent and avirulent Clavibacter michiganensis subsp. michiganensis. Phytopathology. 91:46. [ Links ]
Milijašević, S.; Todorović, B. : Rekanović, E.; Potočnik, I. and balaž, J. 2007. Clavibacter michiganensis subsp. michiganensis, bacterial canker of tomato: 2. Comparison of the effectiveness of extraction procedures and sensitivity of methods for detection in tomato seeds. Pestic. Phytomed. Belgrade, Serbia. 22(2):121-130. [ Links ]
Milijasevic, M. S.; Gartemann, K. H.; Frohwitter, J.; Eichenlaub, R.; Todorovic, B.; Rekanovic, E. and Potocnik, I. 2012. Characterization of Clavibacter michiganensis subsp. michiganensis strains from recent outbreaks of bacterial wilt and canker in Serbia. Eur. J. Plant Pathol. 134(4):697-711. [ Links ]
Normas regionales de la NAPPO (NAPPO). 2013. Organización norteamericana de protección a las plantas sobre medidas fitosanitarias (NRMF):1-8 pp. [ Links ]
Nemeth, J.; Laszlo, E. and Emody, L. (1991). Clavibacter michiganensis ssp. Insidiosus in lucerne seeds. Bull. OEPP 21:713-718. [ Links ]
Pastrick, K. H. and Rainey, R. A. 1999. Identification and differentiation of Clavibacter michiganensis subspespecies by polymerase chain reaction-based techniques. J. Phytopathol. 171:687-693. [ Links ]
Riley, I. T. 1987. Serological relationships between strains of coryneform bacteria responsible for annual ryegrass toxicity and other plants pathogenic corynebacteria. Int. J. Syst. Bacteriol. 37:153-159. [ Links ]
Rodríguez, M. M. 2013. Biogeografía, diagnóstico y manejo integrado de Clavibacter michiganensis subsp. michiganensis en México. Rev. Mex. Fitopatol. 31:32. [ Links ]
SIAP. 2013. Una mirada al panorama agroalimentario de México y el mundo. SAGARPA. 18. http://www.campomexicano.gob.mx/boletinsiap/018-e.html. [ Links ]
Sousa, S. M.; Cruz, L.; Norskov, P. and Rasmussen, O. F. 1997. A rapid and sensitive detection of Clavibacter michiganensis subsp. michiganensis in tomato seeds by polymerase chain reaction. Seed Sci. Technol. 25:581-584. [ Links ]
Zhao, W. J.; Chen, H. Y.; Zhu, S. F.; Xia, M. X. and Tan, T. W. 2007. One-step detection of Clavibacter michiganensis subsp. michiganensis in symptomless tomato seeds using a Taqman probe. J. Plant Pathol. 89(3):349-351. [ Links ]
Received: July 2016; Accepted: September 2016











 texto em
texto em 



