Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Similares em
SciELO
Similares em
SciELO
Compartilhar
Revista mexicana de ciencias agrícolas
versão impressa ISSN 2007-0934
Rev. Mex. Cienc. Agríc vol.7 no.4 Texcoco Mai./Jun. 2016
Articles
Validation of the use of molecular markers of sex and color in hybrids derived from crosses of Maradol x creole papaya
1 Unidad de Biotecnología, Centro de Investigación Científica de Yucatán A.C (CICY). México. (marielavc3@hotmail.com; santey@cicy.mx; gfuentescicy@gmail.com).
2 Unidad de Recursos Naturales, Investigación Científica de Yucatán A. C (CICY). Calle 43 No. 130, Colonia Chuburná de Hidalgo, C. P. 97200, Mérida, Yucatán, México. Tel: (52) 999 942 83 30. (jomijangos@cicy.mx; arimat@cicy.mx).
3INIFAP- Mocochá. Carretera Mérida-Motul, km 25 Mérida C. P. 97454, Mocochá, Yucatán. México. Tel: (01) 991 916 22 15. (zavala.manuel@inifap.gob.mx).
The shape, size and color of fruit pulp are aspects that determine consumer acceptance and priority in the breeding papaya. The objective was to validate the use of molecular markers to select genotypes in terms of sexual type, color and size of the fruit. Genotypes (F1 and F2) derived from intraspecific crosses L7xM22 were characterized molecularly in juvenile stage. To 6 to 9 months compared to classification based on morphological characters. The CPM1815Y52 marker (sexual) was used, the marker CPFC1 (flesh color) as well as eight QTLs associated with fruit size. The CPM1815YC2 marker identified hermaphrodite and female plants in a 2: 1, 100% of F1 and F2 genotypes. With the score CPFC1 identified in the F1 population, 53.57% had orange yellow flesh and 46.43% orange-red pulp. In the F2, the homozygous recessive allele orange-reddish color introgression in 98.1%, 92.6% and 100% over H90B, H70B and H77B. The molecular prediction of floral type and color of pulp, checking phenotypically seedling stage 3 and 7 months before the presence of flowers and fruits is very useful in breeding programs assisted by markers, no QTL studied was able to predict the size or shape of fruit; further research is needed to develop better related to the size and shape of fruits of papaya hybrid between Maradol x creole materials QTLs (or other molecular markers).
Keywords: Carica papaya L.; fruit morphology; molecular markers
La forma, tamaño y color de pulpa del fruto son aspectos que determinan su aceptación por el consumidor y prioritarios en el mejoramiento genético de papaya. El objetivo fue validar el uso de marcadores moleculares para seleccionar genotipos en términos del tipo sexual, color y tamaño del fruto. Genotipos (F1 y F2) derivados de la cruza intraespecífica L7xM22 fueron caracterizados molecularmente en etapa juvenil. Hasta los 6 a 9 meses se comparó con la clasificación basada en caracteres morfológicos. Se utilizó el marcador CPM1815Y52 (tipo sexual), el marcador CPFC1 (color de pulpa), así como ocho QTLs asociados al tamaño del fruto. El marcador CPM1815YC2 identificó plantas hermafroditas y femeninas en una proporción 2:1, en 100% de los genotipos F1 y F2 evaluados. Con el marcador CPFC1 se identificó que en la población F1, el 53.57% presentó pulpa amarilla naranja y 46.43% pulpa naranja-rojizo. En la F2, el alelo homocigoto recesivo de color naranja-rojizo se introgresó en 98.1%, 92.6% y 100% respecto a H90B, H70B y H77B. La predicción molecular del tipo floral y el color de pulpa, al comprobarse fenotípicamente en etapa de plántulas con 3 y 7 meses de anticipación a la presencia de flores y frutos es de gran utilidad en programas de mejoramiento asistido por marcadores, Ningún QTL estudiado, fue capaz de predecir el tamaño, ni la forma de fruto; investigaciones posteriores serán necesarias para desarrollar QTLs (u otros marcadores moleculares) mejor relacionados al tamaño y forma de frutos de papaya de híbridos entre Maradol x material criollo.
Palabras clave: Carica papaya L.; marcadores moleculares; morfología del fruto
Introduction
The species Carica papaya L. is believed to be native to southern Mexico and neighboring Central American countries; currently, it is distributed through tropical and subtropical regions of the world (Fuentes and Santamaria, 2014). Although Mexico is one of the leading producers of papaya, it does not have local varieties. The Maradol variety is widely used, which has demerited quality shape, size (Posada et al., 2010) and the color of the fruit pulp. The shape of papaya fruit depends on the variety and type of flower which has formed. Usually female flowers generate round or spherical to oval fruits, while the hermaphrodite f lowers produce elongated, cylindrical, obovoid or pirifotrmes fruits (Magdalita and Mercado, 2003).
In several countries elongated fruit have greater market acceptance (Manica, 1996; Alonso et al., 2008). In Latin America there is a strong preference for large papaya fruit in domestic markets (Ferraguetti, 2003). According to Marin et al. (2003) there is a preference for hermaphrodite papaya plants with pear-shaped, elongated format, which is associated with a lower ovarian cavity and thicker pulp, a feature that gives them greater commercial value to this kind of fruit in the market. Therefore, it is interesting to know the sex of the parent to obtain a new material cultivated with auspicious segregation hermaphroditic plants that favor the presence of these fruits. The phenotypic sex in papaya prediction is difficult in the early stages of plant development (Sanchez-Betancourt and Nuñez, 2008). The place three plants bowl and schedule a thinning three months after transplantation, favors the presence of at least one hermaphrodite plant to produce elongated fruit, however, expenditures increase for crop management (Kanchana-udomkan et al., 2014).
The DNA markers such as SCAR, RAPD, RFLP and DAF have been associated with sexual type in papaya plants (Deputy et al., 2002; Urasaky et al., 2002; Lemos et al., 2002; Chaves and Nuñez, 2007; Somsri and Bussabakornkul, 2008). Meanwhile the color of the fruit depends on the composition of carotenoids. Only in Hawaiian papayas red pulp, Yamamoto (1964) showed that red fruits accumulate lycopene representing 63% of total carotenoid content. Meanwhile yellow fruits owe their color to an accumulation of β-carotene and β-cryptoxanthin, which represents 75% of total carotenoid content. While red fruit pulp contains about half of that amount. To determine the color of the fruit pulp that have a certain variety by phenotypic methods have to wait for the plant to produce its first fruit months later. In the first fruits papaya are emitted approximately 7 months after transplantation (Calixto, 2011).
Between 7 to 9 months after transplantation, when the first fruits are generated. In relation to fruit size, you can see a wide variation in the size of papaya fruit. We recorded length ranges 7-30 cm (Asudi et al., 2010) and fruit weight from less than 100 g to 10 kg (Jimenez et al., 2014). However, commercial fruits have a weight range of 0.5-2.0 kg and a length of fruit from 10 to 20 cm (Samson, 1986). Demey et al. (2003) and Olmos (2004) suggest that the identification of DNA sequences close to the gene or locus of agronomic interest can be used as a tool to accelerate the processes of traditional crop selection and facilitate the work of plant breeders. Blas et al. (2010) developed a molecular marker (CPFC1) associated with the color pulp Carica papaya L. Hawaiian type and Chen et al. (2007) reported a marker that identifies hermaphrodite and female plants in papaya Sunup and AU9.
In this research, by validating molecular markers is to determine at an early age, sexual type of the plant as well as the color and size of fruit to encourage early selection of promising materials without having to wait for materials to produce f lowers and fruits, with the imminent reduction in time to generate new varieties. For this, molecular markers for sexual type, color pulp eight QTLs associated with morphological characteristics of the fruit were used in a segregating population of papaya derived from the cross between a wild genotype x commercial one (var. Maradol). A young plants of both the parent and the materials derived from the cross, they extracted DNA and classified based on the results based on molecular markers analysis, months later, the grouping based on these markers, was collated with the grouping based on visual and morphological characteristics of flowers and fruits of these plants.
Materials and methods
Vegetal material
The experiment was conducted in two periods: october 2009 to may 2010 and september 2012 to april 2013, the Scientific Research Center of Yucatán A.C. (CICY) in Merida,Yucatan, Mexico. The average annual temperature of the experimental site is 26.1 °C and precipitation 1.012 mm (Conagua). From the cross between creole papaya line 7 (hermaphrodite, pulp yellow-orange) and papaya Maradol 22 (hermaphrodite, orange-red pulp), the 42 seedlings F1 were developed and self-pollinations of H90B, H77B and H70B (genotypes F1) were performed for the development of 162 seedlings F2 of which 83% of the population was ruled out for evaluation by the attack of a viral illness. This research was conducted opencast in a nursery with perimeter fence. The plants were irrigated handled with proper fertilization and pest and weed control following the technology package described in (Cituk, 2006). The parents are homogeneous and stable materials.
Molecular characterization
The leaf material F1 and F2 seedlings was used and a method of DNA extraction based was used on CTAB suggested by Doyle and Doyle(1990), with some modifications. They were amplified by PCR molecular markers CPM1815YC2 (Chen et al., 2007) and CPFC1 (Blas et al., 2010) associated with the character of sex and color of pulp, respectively; QTLs associated markers addition to fruit morphology such as P3K1700CC, ctg-43C0, CPM1556C0 and P3K4311bK0 for weight P3K6372CC and P6K969CC for length, CPM1550C0 for diameter and P3K2426aK3 to form (Blas, 2008). The fragments were visualized by agarose electrophoresis 5%. The expected fragment size was calculated in silico with prfectBLAST software, which indicated the approximate migration of the amplified for molecular markers in parental (L7 and M22) and progeny F1 and F2 product.
Morphological characterization
Throughout the F1 progeny and only 17% of population F2 three fruits per plant were harvested at physiological maturity, this maturity was determined according to Santamaria et al. (2009). The fruits were transported to the laboratory of Molecular Physiology CICY and weight were evaluated, the length, diameter and shape and color of the fruit-based descriptors reported by the UPOV (2010).
Results and discussion
Molecular analysis
The molecular markers linked to QTL characters of commercial interest (quantitative trait loci for its acronym in English) are a tool to increase the efficiency of traditional breeding programs; as they serve to identify, select and maintain genotypes containing the combination of desired alleles and to discard those that do not occur; allowing a decrease in demand for time and resources in studies aimed at the selection of elite genotypes (Valadez and Gunter, 2000).
Molecular marker CPM1815YC2: sex identification Carica papaya L.
The size of the expected fragment molecular marker CPM1815YC2 was predicted bioinformatic program by prfectBLAST (Santiago and Ramirez, 2012) was 167 bp. This molecular marker linked to the sexual nature of Carica papaya L. amplified a double band or single-band, depending on the type of plant analyzed. The amplicon size genotypes dual band corresponds to 175 and 195 bp respectively, while in genotypes with a single band of 190 bp (Figure 1a). The creole parental Line 7 and Maradol 22 had double amplified band, as both are hermaphroditic plants. Similarly, double band appeared in 28 genotypes F1, while the remaining 14 individuals F1 plants corresponding to one female (Figure 2a) band was observed. In the F2 progeny of self-pollination generated H90B, H77B and H70B showed 35, 40 and 39 genotypes dual-band, and 19, 14 and 15 genotypes with a single amplified band, respectively (Figure 3a).
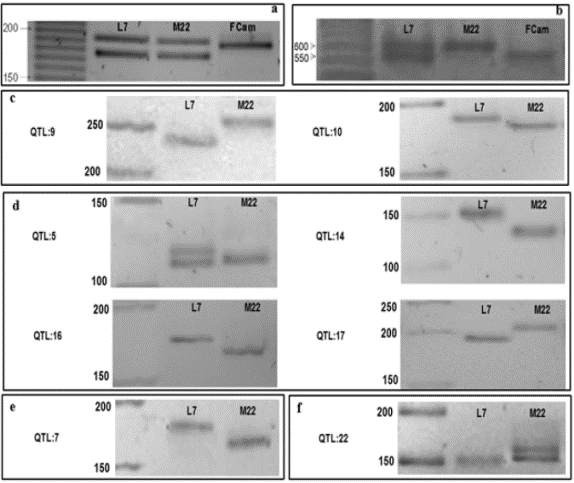
Figure 1 Electrophoresis molecular marker sex CPM1815YC2 (a) and colored pulp CPFC1 (b) amplified parental Creole line 7, Maradol 22 and a female wild plant and light yellow pulp Carica papaya L., and eight QTLs associated length (c), weight (d), diameter (e) and form (f) of the fruit, agarose gel high resolution 5% stained with ethidium bromide. Molecular weight marker 10 bp and 50 bp.
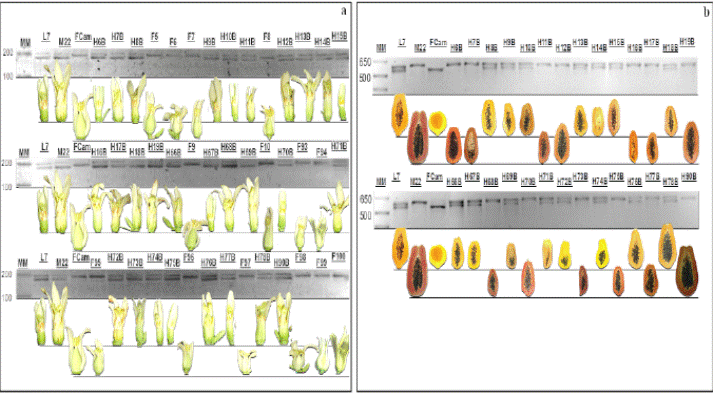
Figure 2 Electrophoresis molecular marker sex CPM1815YC2 (a) and colored pulp CPFC1 (b) parental Criolla amplified L7, Maradol 22 and F1 progeny derived from the cross of Carica papaya L. L7xM22 L7: population Creole line 7, M22: Maradol 22, CFFM: wild female, H: hermaphroditic plants agarose gel 5% ethidium bromide stained. MM: 1 Kb. The photography is included flower and fruit for each F1 progeny obtained at 6 and 9 months after taking the DNA sample, respectively.
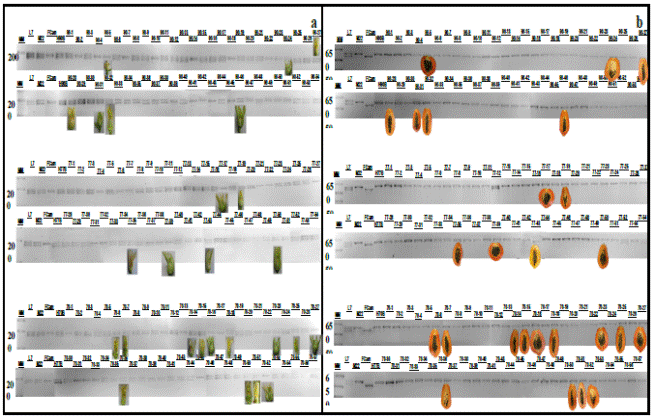
Figure 3 Electrophoresis of molecular marker sex CPM1815YC2 (a) and colored pulp CPFC1 (b) parental Creole amplified L7, Maradol 22 F2 progeny from selfing of H90B, H77B and H70B in Carica papaya L. L7: population Creole line 7, M22: Maradol 22, CFFM: wild female, H: hermaphroditic plants agarose gel 5% ethidium bromide stained. MM: 1Kb. The photography flower and fruit for each progeny F2 obtained at 9 months after taking the DNA sample is included.
These results agree with those reported by Chen et al. (2007) who tested this molecular marker in Hawaiian papaya materials and identified as female and hermaphrodite genotype AU9 and Sunup, respectively with a double band respectively. Our data show that 42 plants in the F1 progeny are hermaphrodites 66.6% and 33.3% are female (ratio 2: 1; hermaphrodites: female). This relationship is expected in an allelic segregation of a selfed hermaphrodite plant or both parents herms as reported by Ming et al. (2007); Niroshini, et al. (2008).
While in the F2 progeny derived from self-pollination of H70B, H77B and H90B presented on the one hand 72.2%, 74.4% and 64.8% of allelic condition hermaphrodite, and 27.8%, 25.9% and 35.2% female, respectively allelic condition. These results are consistent proportionately with those reported by Esquivel et al. (2008) for HGxMR cultivars (65.5 and 34.5%, hermaphrodite and female, respectively), HGxMA (57.8 and 42.2%, hermaphrodite and female, respectively) and Red Maradol (68.9 and 31.04%).
Sexual Carica papaya L. type can only be displayed until started flowering (from three months after transplant approximately) as reported Ming et al. (2007); Niroshini, et al. (2008); Reddy et al. (2012). So determining the sex of papaya seedlings at the molecular level before transplantation would be an advantage in saving time, space and money for the breeder, being a complement to the traditional selection of suitable plants for fruit production agreement with market demands or to incorporate them into the program selection and breeding of the species (Sánchez-Betancourt and Nuñez, 2008).
In addition, papaya fruits from hermaphrodite flowers are generally elongated, with sufficient strength to withstand the mechanical post-harvest and higher commercial demand for occupying less space per unit volume at the time of packaging damage, representing savings freight; mainly for export markets (Muñozcano and Martinez, 2009). The validation of the marker CPM1815YC2 to discriminate female plants hermaphroditic in 42 genotypes F1 (from L7xM22) and 162 genotypes F2 derived from self-pollination of H70B, H77B and H90B is reliable because it was achieved at the molecular level to identify and classify 100% both sex types in papaya plants which shows great potential for use in marker-assisted selection (SAM) in other genotypes wild, creoles and commercial.
Molecular marker CPFC1: color identification Carica papaya L.
The coloring papaya pulp, besides the size and shape of the fruit is an aspect that determines its acceptance by the consumer. From a nutritional standpoint, identifying and obtaining materials aimed at obtaining Carica papaya L. with orange-red pulp favors the presence of a higher content of lycopene, which gives it the orange-red color to the fruit pulp. The intensity of the color depends on the concentration of this pigment, which acts as antioxidants to benefit human health by reducing the risk of lung, stomach and prostate (Giovannucci, 1999) cancer. While in genotypes yellow fruit pulp, the content of β-carotene is higher than the content of lycopene. With the color marker pulp (CPFC1) three allelic forms were observed in the progeny evaluated.
The parental M22 whose phenotypic characteristic was having reddish-orange pulp and 13 genotypes F1 showed a single band with size of 600 bp, which is homozygous recessive condition allelic. While the parental L7, which has yellow flesh and 15 individuals F1 dual band presented a 600-bp and 550 bp another characteristic allelic heterozygous genotypes. In addition, a wild material (FCam) was analyzed with color from light yellow flesh and a band of 550 bp was observed; that is, its condition is homozygous dominant allele (Figure 1b and 2b), these data were close to 568 bp calculated by prfectBLAST.
At the molecular level, the ratio F1 population was approximately 53.57% for genotypes with yellow pulp, and 46.43% for genotypes with reddish-orange pulp (ratio 1:1 approximately) (Figure 2b). Within the F2 progeny tested was observed that the recessive allele of orange-red color introgression 100% for F2 progeny from genotype H90B, in 98.1% of the genotypes from H70B and 92.6% of the genotypes from H77B (Figure 3b). Significantly, in the F2 population from H70B, heterozygous genotype H70-57 presented segregating parental condition L7, moreover, in the F2 population from H77B, the genotypes H77-12, H77-52, and H77- 54, they presented segregating parental heterozygous condition L7 and only genotype homozygous dominant H77-43 presented condition similar to that presented the FCam.
Allelic condition obtained with the color marker CPFC1 in F1 and F2 progeny of this research coincided with the reporting Blas et al. (2010), who indicate that the material SunUp color red pulp amplifies a single band and has homozygous recessive condition, while Richter and Rainbow hybrids show yellow flesh and are heterozygous condition; i.e. contain two alleles showing a double amplification bands. They also mentioned that the Kapoho material has yellow flesh color but has a dominant homozygous condition. In this regard, Rodriguez (2008) reported that the characteristic yellow-fleshed (dominant allele) predominates over the orange-red flesh, so in wild populations rarely papaya appear orange-red fruits, because this color is given by recessive genes.
The molecular marker flesh color (CPFC1) the F1 and F2 genotypes derived from the cross L7xM22 facilitated early detection of genotypes with reddish-orange pulp (phenotypically could be obtained 7-9 months after transplantation) and achieved a correlation molecular much as morphological in relation to the expected segregation ratio. So this specific marker can also have an important potential use in marker-assisted selection (SAM) for the character color of pulp, used in early in nursery plants without having to wait up to nine months to display color fruit of the genotypes used in breeding programs papaya.
QTLs associated markers fruit morphology of Carica papaya L.
The sizes of fragments obtained in silico with values between 84 bp to 174 bp (data not shown) it was as expected and approximate the QTLs microsatellite (SSR) amplified by PCR in parental L7, M22 and progeny F1 and F2 (Figure 4) also consistent with those reported by Mittal and Dubey (2009) who mentioned that the amplified fragments of SSR are variant size, usually several tens to several hundred base pairs (bp), codominant, polymorphic and Mendelian segregation, attributes that could be seen in this investigation.
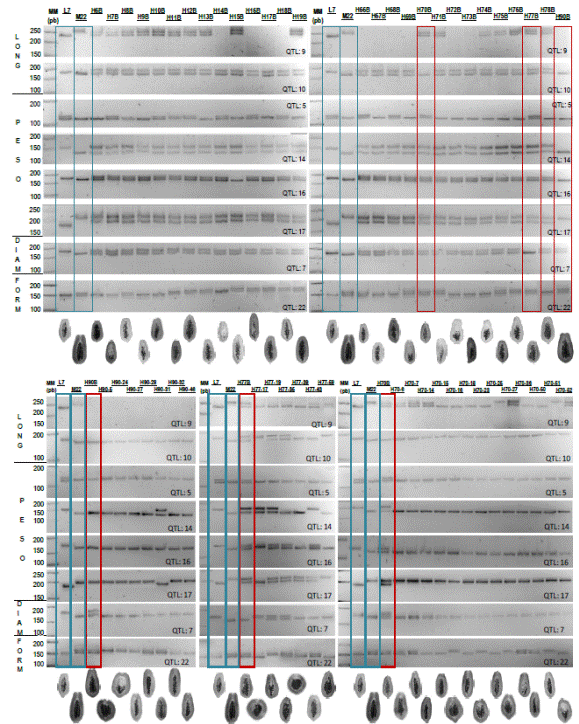
Figure 4 Electrophoresis is an of molecular markers associated with QTLs fruit morphology amplified in line 7 and Maradol 22 Criolla Carica papaya L. parental and progeny F1 (a) and F2 (b). The photography fruit includes corresponding each progeny F1 and F2, obtained 9 months after taking the DNA sample. MM: 50bp.
The eight QTLs associated characters morphology fruit parental L7 and M22, are shown in Figure (1 cf) 2 associated length (1c), 4 associated with weight (1d), one associated diameter (e) and 1 to form (f) of the fruit. None of the eight QTLs or parental and F1 and F2 progeny showed molecular and phenotypic correlation (Figure 4, Table 1); i.e. not provide useful for use in marker-assisted selection for papaya fruit characteristics such as length, weight, diameter and shape of fruit information. It is possible that the lack of correlation is because these characters exhibit a wide variation along a continuous gradient attributed to the interaction of two or more genes (polygenic) (Obando et al., 2008). It is not excluded that there pleiotropic effects on QTLs associated with papaya fruit morphology, which are closely related to the genes ovate, sun and fw.2.2 present in the genome of tomato and orthologous regions in the genome of papaya (Paull et al., 2008; Blas et al., 2012.
Table 1 Mean values associated morphological characters the size and shape of fruit progeny F1 and F2 intraspecific crosses from Carica papaya L. L7xM22.
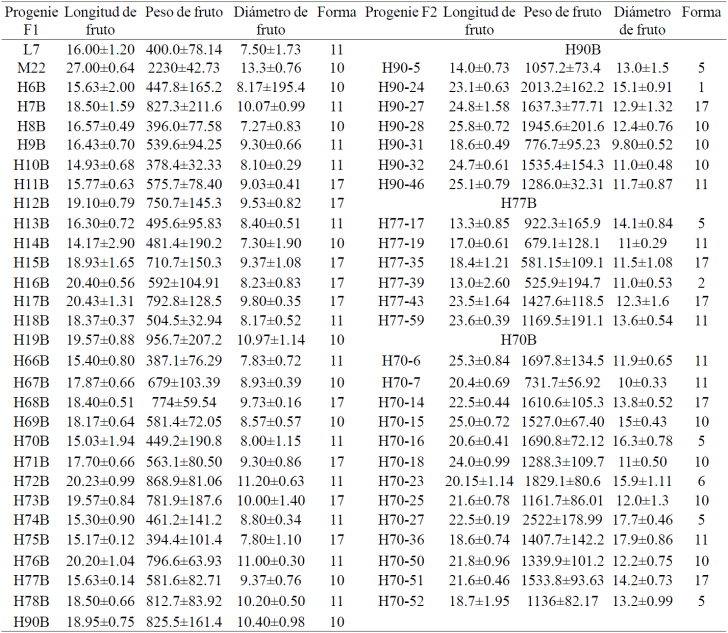
In other species such as tomato, it has been suggested that factors such as fertilization, temperature and humidity will directly influence the expression of the sun gene regulating early elongation pericarp ovary, ovate gene involved in elongation pericarp ovary after fertilization and fw2.2 gene that regulates cell division and fruit development. It is assumed that the effect may be similar papaya, so that the QTL that are influenced by these genes will be affected, such is the case of some genotypes. Or, these QTLs initially evaluated in Hawaiian papaya, are not associated with morphological characteristics of the fruit such as length, weight, diameter and shape of the fruit in hybrids Maradol x creole material Carica papaya L.
Morphological analysis
The molecular identification of sexual coincided with phenotypic data recorded and evaluated three months after transplantation, both parental and F1 and F2 of Carica papaya L. (Figures 2a and 3a) progeny. The F1 population obtained from intraspecific crosses L7xM2 and F2 from self-pollinations of H90B, H77B and H70B genotypes developed only hermaphrodite and female plants. For F1 values were 28 hermaphrodites and 14 female; it is representing 67% of hermaphroditism and 33% of feminism. Within the F2 percentages were 64%, 72% and 67% of hermaphroditism and 36%, 28% and 33% of feminism, for H90B, H77B and H70B, respectively, who remained close to the ratio of 2:1 expected.
Similarly the molecular identification of fruit color was compared with the morphological characterization of the color of the fruits made between 7 and 9 months later, since the plants in both parental and progeny F1 and F2 corresponding issued fruits (Figures 2b and 3b). Similarly 7 and 9 months after taking samples for molecular analysis of QTLs associated with fruit shape, variability in the length, diameter, weight and shape of the corresponding fruits it was characterized and shown in Figure 4 and in Table 1.
Conclusions
The marker CPM1815YC2 discriminating marker female plants hermaphroditic in 42 genotypes F1 and F2 genotypes 164 from mattings L7xM22 molecular level before transplantation. This classification coincided 100% with visually made in the same plants 3 months later. Proving that this marker is highly reliable and has great potential for use in marker-assisted selection (SAM) in Carica papaya L. Morphological sexual identification in plants Carica papaya L. from crossbreeding resulted L7xM22 between 67% to 72% of hermaphrodite plants and between 28.4% to 35.8% of female progeny plants in F1 and F2 progeny; 1: that is, an approximate ratio of 2 was obtained. This corresponds to segregation of character sex when artificial pollination is between two individual’s hermaphrodites or self-pollinated as was the case with the L7 and M22 parental and H90B, H77B and H70B genotypes.
The classification as fruit pulp orange-red in both parental and progeny F1 and F2 from the cross L7xM22 at the molecular level in young plants coincided 100% with the morphological analysis in the fruits issued by the same plants between 7 and 9 months after. So the molecular marker pulp color (CPFC1) has a high potential for use in breeding programs assisted by select fruit oriented red markers. However, this marker does not distinguish between colored fruits light yellow pulp, pulp of those orange.
The QTLs associated markers character weight, length, diameter and shape of the papaya fruit even if they could make and showed allelic condition in several of the contrasting characteristics of the parents, were not useful for assisted selection by molecular markers (SAM) of individuals in such contrasting characters in F1 and F2 progeny of hybrids from the intra-specific crosses L7xM22. further research is needed to develop better related to the size and shape of fruits of papaya hybrid between Maradol x creole materials QTLs (or other molecular markers). However, it is possible that the QTLs tested here can distinguish fruit shape in other materials Carica papaya L.
Literatura citada
Alonso, E. M.; Tornet, Q. Y.; Ramos, R. R.; Farrés, A. E.; Aranguren, G. M. y Rodríguez, M. D. 2008. Caracterización y evaluación de dos híbridos de papaya en Cuba. Agric. Téc. Méx. 34(3):333-339. [ Links ]
Asudi, G. O.; Ombwara, F. K.; Rimberia, F. K.; Nyende, A. B.; Ateka, E. M.; Wamocho, L. S.; Shitanda, D. and Onyango, A. 2010. Morphological diversity of Kenia papaya germplasm. Afr. J. Biotechnol. 9(51):8754-8762. [ Links ]
Blas, A. L. 2008. Molecular genetic basis of papaya fruit traits. Tesis de doctorado. University of Hawaii. Hawaii, USA. 185 p. [ Links ]
Blas, A. L.; Yu, Q.; Veatch, O. J.; Robert, V.; Paull, H. M. and Ming, R. 2012. Genetic mapping of quantitative trait loci controlling fruit size and shape in papaya. Mol. Breed. 29(2):457-466. [ Links ]
Blas, A.; Ming, R.; Liu, Z.; Veatch, O. J.; Robert, E.; Paull, H. and Qingyi, Y. 2010. Clon in g of the papaya chromoplast-specificly copene β-Cyclase, controlling fruit flesh color reveals conserved microsynteny and a recombination Hot Spot. Plant Physiol. 152(4):2013-2022. [ Links ]
Chaves, B. G. and Nuñez, V. 2007. A SCAR marker for the sex types determination in Colombia genotypes of Carica papaya L. Euphytica 153(1):215-220. [ Links ]
Chen, C.; Yu, Q.; Hou, S.; Li, Y.; Eustice, M.; Skelton, R. L.; Veatch, O.; Herdes, R. E.; Diebold, L.; Saw, J.; Feng, Y.; Qian, W.; Bynum, L.; Wang, L.; Moore, P. H.; Paull, R. E.; Alam, M. and Ming, R. 2007. Construction of a sequence tagged high density genetic map of papaya for comparatative structural and evolutionary genomics in brassicales. Genetics. 177(4):2481-2491. [ Links ]
Cituk-Chan, D. E. 2006. Paquete tecnológico de producción de papaya Carica papaya L. Instituto Tecnológico de Conkal. 62 pp. [ Links ]
Demey, J. R.; Zambrano, Y. A.; Fuenmayor, F. y Segovia, V. 2003. Relación entre caracterizaciones molecular y morfológica de una colección de yuca. Interciencia. 28(12):684-689. [ Links ]
Doyle, J. J. and Doyle, J. L. 1990. A rapid total DNA preparation procedure for fresh plant tissue. Focus 12:13-15. [ Links ]
Esquivel, M.; Tornet, Y.; Ramos, R. E.; Farrés, E.; Castro, J. y Rodríguez, M. C. 2008. Evaluación de tres cultivares de papaya del grupo ‘SOLO’ basada en caracteres de crecimiento y productividad. Cultivos Tropicales. 29(2):59-64. [ Links ]
Ferraguetti, G. A. 2003. Caliman 01- O primeio híbrido de mamão mamão Formosa Brasileiro. In: Martins, D. S. (ed.). Papaya Brasil: qualidade do mamão para mercado interno. Incaper. Vitoria, ES. 211-218 pp. [ Links ]
Fuentes, G. and Santamaría, J. M. 2014. Chapter 1: Papaya (Carica papaya L.): origin, domestication, and production. Genetics and genomics of papaya. Plant genetics and genomics: crops and models. Spinger. 10: 3-15. [ Links ]
Giovannucci, E. 1999. Tomatoes, tomato-based products, lycopene and cancer: review of the epidemiologic literature. Journal of the National. Cancer Institute. 91(4):317-331. [ Links ]
IBPGR. 1988. Descriptors for papaya. Bio. Int. Roma, Italia. 34 pp. [ Links ]
Jimenez, V. M.; Mora, N. E. and Gutierrez, S. M. V. 2014. Plant genetics and genomics, genetics and genomics of papaya: chapter 2: biology of the papaya plant. Crops and Models. Spinger. 10:17-33. [ Links ]
Kanchana-udomkan, C.; Ford, R. and Drew, R. 2014. Plant Genetics and Genomics, Genetics and genomics of papaya, chapter 19: molecular markers in papaya. Crops and Models. Spinger. 10:355-375. [ Links ]
Lemos, E. G. M.; Silva, C. L. S. P. and Zaidan, H. A. 2002. Identifacation of sex in Carica papaya L. using RAPD markers. Euphytica. 127(2):179-184. [ Links ]
Magdalita, P. M. and Mercado, Ch. P. 2003. Determining the sex of papaya for improved production. Food and fertilizer technology center. FFTC publication database, extension bulletins 534. University of the Philippines at Los Banos College. Laguna, Philippines. 6 p. [ Links ]
Manica, I. 1996. Cultivares e melhoramento do mamoeiro. In: Mendes, L. G.; Dantas, J. L. L.; Morales, C. F. G. Mamão no Brasil. Embrapa Mandioca e Fruticultura. Cruz das Almas, Brazil. 121-143 pp. [ Links ]
Ming, R.; Yu, Q. and Moore, P. H. 2007. Sex determination in papaya. Seminers in cell and developmental biology. 18(3):401-408. [ Links ]
Mittal, N. and Dubey, A. K. 2009. Microsatellite markers- a new practice of DNA based markers in molecular genetics. Pharmacognosy Reviews. 3(6):235-246. [ Links ]
Muñozcano, R. M. y Martínez, A. C. O. 2009. Paquete tecnológico para la producción de papaya en Sinaloa. Fundación Produce Sinaloa A.C. Sinaloa, Mexico. 37 p. [ Links ]
Nagata, M. and Yamashita, I. 1992. Simple method for simultaneous determination of chlorophyll and carotenoides in tomato fruit. J. Japanese Soc. Food Sci. Technol. 39(10):925-928. [ Links ]
Niroshini, E. J.; Everard, M. D. T.; Karunanayake, E. H. and Tirimanne, T. L. S. 2008. Detection of sequence characterized amplified region (SCAR) markers linked to sex expression in Carica papaya L. J. Nat. Sci. Foundation Sri Lanka. 36(2):145-150. [ Links ]
Obando, J.; Fernández, T. J. P.; Martínez, J. A.; Alarcón, A. L.; Eduardo, I.; Arús, P. and Monforte, A. J. 2008. Identification of melon fruit quality quantitative trait loci using near-isogenic lines. J. Am. Soc. Hort. Sci. 133(1):139-151. [ Links ]
OECD. 2005. Consensus document on the biology of papaya (Carica Papaya L.). ECD environment, health and safety publications series on harmonisation of regulatory oversight in biotechnology no. 33. Paris. 66 p. [ Links ]
Olmos, S. 2004. Selección asistida por marcadores moleculares y su aplicación en el mejoramiento genético de trigo. Agrotecnia. 12:23-31. [ Links ]
Paull, R. E.; Irikura, B.; Wu, P.; Turano, H.; Chen, N. J.; Blas, A.; Fellman, J. K.; Gschwend, A. R.; Wai, C. M.; Yu, Q.; Presting, G.; Alam, M. and Ming, R. 2008. Fruit development, ripening and quality related genes in the papaya genome. Tropical Plant Biol. 1(3- 4):246-277. [ Links ]
Posada, P. L.; Gómez, K. R.; Pérez, P. J.; Reyes, V. M. and Norman, M. O. 2010. Development of a new papaya (Carica papaya L.) hybrid IBP 42-99. Interciencia. 35(6):461-465. [ Links ]
Reddy, S. R.; Balamurali, K. R. and Jagadeeswara, R. K. 2012. Sex determination of Papaya (Carica papaya) at seedling stage through RAPD markers. Res. Biotechnol. 3(1):21-28. [ Links ]
Rodríguez, M. A. 2008. De la ciencia popular a la industria: la variedad cubana de papaya “Maradol”. Instituto de Investigaciones Fundamentales en Agricultura Tropical (INIFAT). La Habana, Cuba. 23 p. [ Links ]
Samson, J. A. 1986. Mango, avocado and papaya. In: tropical fruits. JA Samson (Ed). Longman Scientific Inc. New York. 185-221 pp. [ Links ]
Sánchez-Betancourt, S. E. y Nuñez, Z. V. M. 2008. Evaluación de marcadores moleculares tipo SCAR para determinar sexo en plantas de papaya (Carica papaya L.). Rev. Corpoica-Ciencia y Tecnología Agropecuaria. 9(2):31-36. [ Links ]
Santamaría, B. F.; Sauri, D. E.; Espadas, Gil, F.; Díaz, P. R.; Larqué, S. A. and Santamaría, F. J. 2009. Postharvest ripening and maturity indices for Maradol papaya. Interciencia. 34(8):583-588. [ Links ]
Santiago, S. P. y Ramírez, P. J. H. 2012. prfectBLAST: a platform- independent portable front end for the command terminal BLAST+ stand-alone suite. BioTechniques. 53(5):299-300. [ Links ]
Somsri, S. and Bussabakornkul, S. 2008. Identification of certain papaya cultivar sand sex identification in papaya by DNA amplification fingerprinting (DAF). Acta Hortic. 787:197-206. [ Links ]
UPOV. 2010. International union for the protection of new varieties of plants. Papaya UPOV Code: Caric_pap Carica papaya L. 32 p. [ Links ]
Urasaki, N.; Tarora, K.; Uehara, T.; Chinen, I.; Terauchi, R. and Tokumoto, K. 2002. Rapid and highly reliable sex diagnostic PCR assay for papaya (Carica papaya L.). Breed. Sci. 52(4):333-335. [ Links ]
Valadez, M. E. y Günter, K. 2000. Huellas de ADN en genomas de plantas. Teoría y protocolos de laboratorio. Mundi-Prensa México. D. F., México. 60 p. [ Links ]
Yamamoto, H. Y. 1964. Comparison of the carotenoids in yellow-and red-fleshed Carica papaya. Nature 201:1049-1050. [ Links ]
Received: December 2015; Accepted: March 2016











 texto em
texto em 


