Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Revista mexicana de ciencias agrícolas
versión impresa ISSN 2007-0934
Rev. Mex. Cienc. Agríc vol.5 spe 8 Texcoco 2014
Investigation notes
A new strain of Chino del Tomate Virus isolated from soybean plants (Glycine max L.) in Mexico
1Instituto Potosino de Investigación Científica y Tecnológica, San Luis Potosí, S. L. P. 78216, México. (grarguel@ipicyt.edu.mx; berbanher@gmail.com; sambriz@ipicyt.edu.mx).
2Unidad Académica de Agronomía-Universidad Autónoma de Zacatecas, A. P. 336, 98000. Zacatecas, Zacatecas. México.
3Campo Experimental Zacatecas-INIFAP, Calera de V. R., Zacatecas, C. P. 98500, México. (fitovalle58@yahoo.com.mx).
4CIIDIR-IPN, Unidad Sinaloa. P. O. Box 280, Guasave, Sinaloa 81101, México. (jmendezl@ipn.mx).
Molecular techniques such as PCR and RFLP allowed the isolation and identification of complete A component sequences of two begomoviruses from soybean plants with virus symptoms in the state of Sinaloa, Mexico. Both genomes were analyzed by comparison of their nucleotide sequences with those available in the NCBI database. The first isolate corresponded to Rhynchosia golden mosaic virus (RhGMV), while the second corresponded to the Chino del tomate virus (CdTV) which showed a 92.3% overall identity with its closest relatives, CdTV-[8[ and CdTV-[RK[. Plant extracts of symptomatic soybean were analyzed by PCR using a pair of universal primers that allowed to amplify and confirm the presence of the CdTV B component, therefore the second isolate identified in this work is proposed as a new CdTV strain called "Chino del tomate virus - Soybean" (Mexico: Sinaloa: 2005). The presence of this new CdTV strain isolated from soybean is important because to date virus infection was naturally restricted to plants of the Solanaceae family.
Keywords: geminivirus; mixed infection; soybean; RFLP; CdTV
Mediante el uso de técnicas moleculares como la PCR y los RFLPS se logró aislar e identificar a partir de plantas de soya con síntomas de virosis las secuencias completas del componente A de dos begomovirus en el estado de Sinaloa, México. Ambos genomas fueron analizados mediante la comparación de sus secuencias nucleotídicas con las disponibles en la base de datos del NCBI. El primer aislado correspondió a Rhynchosia golden mosaic virus (RhGMV), mientras que el segundo corresponde a la especie Chino del tomato virus (CdTV) el cuál mostró 92.3% de identidad global con sus parientes más cercanos, CdTV-[8] y CdTV-[RK]. Los extractos de plantas de soya sintomáticas fueron analizados mediante PCR utilizando un par de iniciadores universales que permitieron amplificar y confirmar la presencia del componente B de CdTV, por lo que se propone al segundo aislado identificado en este trabajo como una nueva cepa de CdTV denominada "Chino del tomato virus- Soybean'" (México: Sinaloa: 2005). La presencia de esta nueva cepa de CdTV aislada de soya es importante ya que hasta la fecha la infección del virus había sido restringida en forma natural a plantas de la familia Solanaceae.
Palabras clave: geminivirus; infección mixta; soya; RFLP; CdTV
Introduction
Geminiviruses are a diverse and widely distributed group of plant pathogens that cause great economic losses worldwide. Their genome consists of one or two molecules of single-stranded circular DNA with a size of 2.5 to 3 Kb (Lazarowitz, 1992). They are transmitted by insects of the Homoptera order (whiteflies and leafhoppers) to a variety of mono and dicotyledonous plants, producing symptoms such as mosaics, speckled, striped, leaf deformation, dwarfing, yellowing and chlorosis becoming one of the most important groups of plant pathogens from an economic point of view because of its high incidence, distribution and severity of symptoms.
Currently the ICTV (International Comittee on Taxonomy of Viruses) recognizes four genera (Mastrevirus, Topocuvirus, Curtovirus and Begomovirus) belonging to the Geminiviridae family (Fauquet et al., 2008), begomoviruses are the most diversified group with highest worldwide distribution, transmitted by the whitefly (Bemisia tabaci Genn.), their genome can be mono or bipartite and infect dicotyledonous plants. The A and B components of the bipartite begomoviruses share a common region ranging from 160 to 230 base pairs (bp), containing the cis elements required for viral replication (Arguello et al., 1994). The A component produces the proteins involved in replication and encapsidation of the viral genome (Gutiérrez et al. , 2004) while the B component encodes two proteins promoting virus movement through the plant (Gafni and Epel, 2002).
At the beginning of the 70s the first report was published on the presence of a new disease causing crinkle in tomato plants grown in the state of Sinaloa, Mexico (González and Cervantes, 1973). The disease was associated with the presence of large whiteflies populations and it was not until 1984 that it was possible to isolate and characterize the Chino del tomate virus (Brown and Hine, 1984), a begomovirus considered as the causative agent of the disease. Currently CdTV is believed to be restricted to members of the Solanaceae family mainly in tomato and chili crops present in the states of Chiapas, Morelos, Sinaloa and Tamaulipas showing a tendency to spread due to its wide host range (Brown and Nelson, 1988; Torres-Pacheco et al., 1996).
In this paper a new CdTV strain isolated from soybean plants (Glycine max L.) is reported in the state of Sinaloa, Mexico and whose genome differences with respect to other strains reported in Solanaceae could favor its presence in soybean crops.
Collection of samples. 12 samples of soybean plants were harvested in commercial plots located in the Guasave municipality, Sinaloa, Mexico showing virus-like symptoms (dwarfism, rolled and yellow leaves).
DNA extraction. A genomic DNA extraction protocol was adopted for all plant tissue samples analyzed. This was based on a modification of the Della Porta's method (1983) using liquid nitrogen and extraction buffer "A" consisting of 100 mM Tris pH 8, 50 mM NaCl pH 8, 50 mM EDTA pH 8 and distilled water.
Amplification of begomovirus DNA present in soybean extracts. In some instances the viral load contained in DNA extracts from plants with possible virus symptoms is reduced thus the Templiphi kit (GE Healthcare) was used to increase the viral DNA concentration by an isothermal reaction catalyzed by the ¿29 bacteriophage DNA polymerase capable of producing micrograms of single stranded circular DNA from picograms.
Polymerase chain reaction (PCR). Viral DNA amplifications were carried out using the universal primers for begomoviruses: pRep-DGR (Méndez et al., 2006), PrCP70-Mot, prRepQGR-rev (De la Torre et al, 2006) and prC889 (Brown et al, 2001). The PCR products comprise the complete A component. The PCR reaction was performed using as positive control the A genomes of Pepper huastec yellow vein virus (PHYVV) and Pepper golden mosaic virus (PepGMV) cloned into the pGEM-T easy vector (Promega, Madison, WI).
Subsequently the presence of the CdTV-soybean B component was confirmed using the universal primers: BC1-80rev (AGAYGARTATCARYTDTCNCATGA) and BV1-310for (AGGDACNGTN AAGRTYGARCGTGT) designed in this work, which are complementary to sequences of the begomovirus B component from both the new and the old World. For all oligonucleotides sets, the composition of the PCR reaction mixture (50 (L total volume) was the same and consisted of the following: 1X Taq DNA polymerase buffer, 1.5 mM MgCl2, 0.2 mM of each dNTP, 1 μM oligonucleotides, 2.5 U Taq DNA polymerase, 1 (ig DNA. The conditions for viral DNA amplification were: initial denaturation at 94 °C/2min, and 35 cycles of 94 °C/1min, 55 °C/1min, 72 °C/1min, with a final extension of 72 °C/5 min. The amplified products were analyzed by electrophoretic mobility in 1% agarose gels.
Characterization of amplified fragments and clones by restriction pattern (RFLP). Positive clones were selected for each oligonucleotide combination used and were analyzed by RFLP to identify polymorphic bands that indicate whether it is a single or mixed infection. The MspI and HhaI restriction enzymes were used in combination with EcoRI (New England BioLabsTM). The digested products were analyzed by electrophoretic mobility in 2% agarose gels.
Cloning and sequencing
PCR products obtained from field samples were directly cloned into the pGEM-T Easy (Promega, Madison, WI) plasmid, as indicated by the supplier. Transformation of Escherichia coli Top -10 calcium competent cells was performed by the heat shock method. Viral clones obtained were sequenced at the National Laboratory of Agricultural, Medical and Environmental Biotechnology (LANBAMA) in San Luis Potosí, Mexico. The in silico analysis of the nucleotide sequences obtained was performed by comparison with available sequences in the NCBI database, using the BLASTN software and phylogeny analysis was performed using the Clustal V method of MegAlign (DNASTAR software, Lasergene, London).
Begomoviruses identification by PCR
Total DNA extracts obtained from soybean plants analyzed by PCR using universal primers for begomoviruses, prRepDGR, prCP70, prRepDGR-rev and prC889 allowed the amplification from five of the twelve soybean extracts of two overlapping fragments of viral DNA of 1.8 and 1.4 Kb representing the complete begomovirus A component as has been reported previously (Méndez et al., 2006).
PCR amplification of the begomovirus B component
PCR confirmation of the B component corresponding to each A component identified, was obtained in an efficient manner by using the combination of universal primers
BC1-80rev and BV1-310for. This combination of universal primers is complementary to conserved sequences located at begomovirus B components present in the old and new World. The length of the amplified viral B DNA fragments was 1 300 bp.
RFLP analysis
The clones obtained belonging to the begomovirus A and B components were digested with the EcoRI restriction enzyme to confirm that they contained the viral DNA fragments. For the A component the first set of positive clones released a 1.8 Kb fragment amplified with the primer combination prCP70 and prRepDGRrev while the second group of clones released a 1.4 kb fragment corresponding to the viral DNA including the begomovirus intergenic region which was amplified with the universal primer combination prRepDGR and prC889 while for clones obtained from the BC1-80rev and BV1-310for oligonucleotides the presence of a 1 300 bp fragment was confirmed.
Eight positive clones were selected for each oligonucleotide combination used and were analyzed by RFLP. The presence of two different restriction patterns in the clones obtained from the A and B components subjected to RFLP analysis indicated that it was a mixed infection involving two different begomoviruses (Figure 1). Results from clones selection is consistent with the increase in reports of mixed infections by begomoviruses in Mexico, the clearest example is the mixed infections caused by PHYVV and PepGMV, which produce serious damage to chili crops when they co-infect the same plant (Rentería et al., 2011).

Figure 1 Restriction patterns obtained by RFLP from two different clones carrying 1.4 Kb inserts of the A component of the two begomoviruses obtained from soybean plant extracts using the prRepDGR / prC889 oligonucleotide combination. The recombinant plasmids were digested with the EcoRI restriction enzyme in combination with the HinfI enzyme (lines 2 and 3) or MspI (lanes 4 and 5).
The high incidence of mixed infections caused by begomoviruses in weeds and crops of economic interest may influence the emergence of new strains and variants with changes at the nucleotide level in key genes that could produce the appearance of new symptoms, more severe and even extend their natural host range (Brown et al., 2000). Sequence analysis by RFLP is important since it reports the presence of two or more begomovirus genomes infecting the same plant showing that mixed infections are very common in nature (Bañuelos et al., 2012) and allows the identification of small nucleotide differences even among viruses of the same species by the presence of one or more polymorphic bands provided that digestions are performed with enzymes allowing cloned DNA fragment release in combination with another restriction enzyme recognizing cleavage sites of a maximum of five nucleotides, thus revealing more polymorphisms.
The digestion products were subjected to electrophoresis in 2% agarose gel. Lane 1 shows the molecular weight marker (100 bp DNA ladder). The plasmid containing the CdTV-soybean amplicon corresponds to lines 2 and 4 and that corresponding to RhGMV is represented in lines 3 and 5.
Sequence analysis and genomic organization
The sequences obtained were edited using the program edit seq (DNAStar MegAlign software) and the complete genome of a first begomovirus was assembled whose A component (accession number NC010294) is 2604 bp long while the length of the B component (Accession number NC010293) is 2551 bp. The A component of the first isolate from soybean plants was compared to the sequences in the NCBI database and those showing greater similarity at the nucleotide level were analyzed using the Clustal V algorithm of MegAlign and high identity was obtained at nucleotide sequence level (99%) with the A component of the Rhynchosia golden mosaic virus, its close relative. For the second begomovirus a complete A component was obtained (accession number DQ347945) whose 2609 bp sequence was found to be 92.3 % similar to the CdTV-[8] and CdTV [RK] A component, its closest relatives (Table 1). Furthermore, the partial characterization (1300 bp) of the B component (access number EU339940) of the second begomovirus isolated during this work was achieved.
Table 1 Overall comparison (%) between the complete A components of begomoviruses of the CdTV species and their sequence similarity with the CdTV-soybean A component.
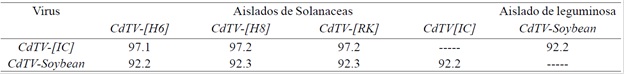
Table 1.The sequence similarity between CdTV-[IC] and CdTV variants isolated from tomato is at least 97%, while CdTV-soybean appears in soybeans and shows a 92.3% similarity with its closest relatives. CdTV-[H6] (access number AF226665), is a variant not mentioned in this report.
Taxonomic criteria established by the ICTV state that comparisons of a begomovirus completeA component with the A component of its closest relative showing differences between 89 and 93%, indicate that the analyzed isolate can be considered a new separate viral strain (Fauquet et al, 2008) therefore, based on the results obtained in the present work the isolation of a new strain of the CdTV species in soybean plants is reported, whose name accepted by the ICTV is Chino del tomato virus- Soybean (Mexico: Sinaloa: 2005), Fauquet et al., 2008.
The two viruses isolated during this study belong to different species and as such have very different iterons (Arguello et al., 1994) thus an interaction between them would not be expected, however, pseudorecombinant formation has been observed between begomoviruses not sharing the same iterons sequences, generating very aggressive infections (Chakraborty et al, 2008) together with the above there is always the possibility that different viruses produce a synergy increasing damages on crops.
This indicates the importance of further analysis on the potential phytosanitary implications of the RhGMV and CdTV presence in soybean crops and the risks that they may propagate into new hosts.
Until now, CdTV had been previously identified naturally infecting only plants of the Solanaceae family. In this paper the first isolation of CdTV-Soybean is reported from a legume therefore emphasizing the need to monitor the incidence of CdTV in soybean crops in order to avoid an outbreak caused by this begomovirus.
Conclusions
Two begomoviruses present in soybean plants collected in the state of Sinaloa, Mexico were characterized at the sequence level. The first isolate was a variant, genetically very close to RhGMV, previously reported in the same geographical area. However, a second isolated begomovirus proved to be a new strain of the CdTV species first isolated from a legume since to date its presence was restricted to Solanaceae. This opens new perspectives regarding these begomoviruses which will provide insights about their ability to infect plants of the Solanaceae family.
Literatura citada
Arguello-Astorga, G. R.; Guevara-González, R. G.; Herrera-Estrella, L. R. and Rivera-Bustamante R. F. 1994. Geminivirus replication origins have a group-specific organization of iterative elements: a model for replication. Virology 203:90-100. [ Links ]
Bañuelos-Hernández, B.; Mauricio-Castillo, J.A.; Cárdenas-Conejo, Y.; Guevara-González, R. G. and Arguello-Astorga, G. R. 2012. A new strain of tomato severe leaf curl virus and a unique variant of tomato yellow leaf curl virus from Mexico. Arch. Virol. 157:1835-1841. [ Links ]
Brown, J. K. and Hine, R. B. 1984. Geminate particles associated with the leaf curl of "chino" disease of tomatoes in coastal areas of western Mexico. Phytopathology 74:844. [ Links ]
Brown, J. K. and Nelson, M. R. 1988. Transmission, host range and virus-vector relationships in chino del tomate virus, a whitefly-transmitted geminivirus from Sinaloa, Mexico. Plant Dis. 72:866-869. [ Links ]
Brown, J. K.; Ostrow, K. M.; Idris, A. M. and Stenger, D. C. 2000. Relationships to other begomoviruses and identification of A-component variants that affect symptom expression. Phytopathology 90:546-552. [ Links ]
Brown, J. K.; Idris, A. M.; Torres, J. I.; Banks, G. K. and Wyatt, S. D. 2001. The core region of the coat protein gene is highly useful for establishing the provisional identification and classification of begomoviruses. Arch. Virol. 146:1581-1598. [ Links ]
Chakraborty, S.; Vanitharani, R.; Chattopadhyay, B. and Fauquet, B. M. 2008. Supervirulent pseudorecombination and asymetric synergism between genomic components of two distinct species of begomovirus associated with severe tomato leaf curl disease in India. J. Gen. Virol. 89:818-828. [ Links ]
De la Torre-Almaraz, R.; Monsalvo-Reyes, A.; Romero-Rodríguez, A.; Argüello-Astorga, G. R. and Ambríz-Granados, S. 2006. A new begomovirus inducing yellow mottle in okra crops in Mexico is related to Sida yellow vein virus. Plant Dis. 90:378. [ Links ]
Dellaporta, S. L.; Wood, J. and Hicks, J. B. 1983. A rapid method for DNA extraction from plant tissue. Plant Mol. Biol. Rep. 1:19-21. [ Links ]
Fauquet, C. M.; Briddon, R. W.; Brown, J. K.; Moriones, E.; Stanley, J.; Zerbini, M. and Zhou, X. 2008. Geminivirus strain demarcation and nomenclature. Arch. Virol. 153:783- 821. [ Links ]
Gafni, Y. and Epel, B. L. 2002. The role of host and viral proteins in intra-and inter-cellular trafficking of geminiviruses. Physiol. Mol. Plant Pathol. 60:231-241. [ Links ]
González, G. R. y Cervantes, R. J. 1973. Enchinamiento de las plantas de tomate, enfermedad en observación y estudio en el Valle de Culiacán. In: Memorias de la Primera reunión sobre plagas y enfermedades en hortalizas en Sinaloa. Secretaría de Agricultura y Ganadería. Culiacán, Sinaloa, México. 51-56. [ Links ]
Gutiérrez, C.; Ramirez-Parra. E.; Mar, C. M.; Sanz-Burgos, A. P.; Luque, A. and Missich, R. 2004. Geminivirus DNA replication and cell cycle interactions. Veterinary Microbiol. 98:111-119. [ Links ]
Lazarowitz, S. G. 1992. Geminiviruses: genome structure and gene function. Critical Reviews in Plant Sciences 11:327-349. [ Links ]
Méndez-Lozano, J.; Perea-Araujo, L. L.; Ruelas-Ayala, R. D.; Leyva-López, N. E.; Mauricio-Castillo, J. A. and Argüello-Astorga, G. R. 2006. A begomovirus isolated from chlorotic and stunted soybean plants in Mexico, is a new strain of Rhynchosia golden mosaic virus. Plant Dis. 90:972. [ Links ]
Ndunguru, J.; Legg, J.; Aveling, T.; Thompson, G. and Fauquet, C. 2005. Molecular biodiversity ofcassava begomoviruses in Tanzania: evolution of cassava geminiviruses in Africa and evidence for East Africa being a center of diversity of cassava geminiviruses. Virol. J. 2:21. [ Links ]
Rentería-Canett, I.; Xoconostle-Cázares, B.; Ruiz-Medrano, R. and Rivera-Bustamante, R. F. 2011. Geminivirus mixed infection on pepper plants: Synergistic interaction between PHYVV and PepGMV. Virol. J. 8:104. [ Links ]
Torres-Pacheco, I.; Garzón-Tiznado, J.A.; Brown, J. K.; Becerra-Flora, A. and Rivera-Bustamante, R. F. 1996. Detection and distribution of geminiviruses in Mexico and the southern United States. Phytopathology 86:1186-1192. [ Links ]
Received: March 2014; Accepted: April 2014











 texto en
texto en 


