Introduction
Hypersaline environments such as salt lakes, hypersaline ponds and saline-alkali soils (Ghai et al. 2011; Shi et al. 2012; El Hidri et al. 2013; Purohit et al. 2014) are distributed worldwide and contain a large number of microorganisms from all domains of life called halophiles, well adapted to these hostile conditions (Yin et al. 2015). Halophiles have been divided in two groups dependent on their tolerance to salt. Halotolerant, when they are able to grow at high salt concentrations but do not require it for growth (Purohit et al. 2014) and extreme halophiles that require high salt concentrations for growth. Most halophilic bacteria belong to the Firmicutes (e.g., Bacillus, Oceanobacillus, Alkalibacillus, Virgibacillus, Halobacillus), Actinobacteria (e.g., Leucobacter, Arthrobacter and Nesterenkonia) or Proteobacteria (i.e., Halomonas, Chromohalobacter and Halovibrio) (Romano et al. 1996; Yumoto et al. 2005; Gonzalez-Domenech et al. 2009; Luo et al. 2009; Whon et al. 2010; Tang et al. 2011; El Hidri et al. 2013; Govender et al. 2013). Halophiles can also be alkaliphiles that grow optimally at pH 9 but are unable to grow at pH 7 or alkalitolerant with an optimal growth at a neutral pH but that can grow at pH 10 (Purohit et al. 2014). Haloalkaliphic microorganisms are both halophilic and alkaliphilic and live in saline alkaline environments widely distributed on earth. They can grow at pH > 10 and salt content up to 25% (w/v) (El Hidri et al. 2013).
There has been an increasing interest in haloalkaliphilic microorganism due to their ability to grow under extreme conditions and their enzymes that are well adapted to high pH and high salt concentration, which can be applied in industrial process. Enzymes from haloalkaliphilic bacteria can be used at high salt concentrations and alkaline pH; conditions found often in detergent, leather or silk industries in which enzymes with poor a lower stability in these conditions can not be used (Abdel-Hamed et al. 2016).
Halophilic and alkaliphilic microorganism can also synthesize products with industrial applications, such as polyhydroxyalkanoates (PHAs) and ectoine. The latter is an ingredient in many cosmetics, skin care products and medicinal preparations, and has been used as an enhancer in molecular biology techniques (Ma et al. 2010). Haloalkaliphilic bacteria produce halophilic hydrolases (proteases, xylanases, cellulases, amylases), bacteriorhodopsin, antimicrobials substances, bio-surfactants or bio-emulsifiers that have many industrial applications. For instance, proteases are applied in detergent, textile, leather, silk industries and have various pharmaceutical applications (Purohit et al. 2014; Yin et al. 2015). Siderophores are organic metal-chelating agents produced by microorganisms especially in Fe-limiting conditions. They can bind to other metals and can be used in biosensors, and as biocontrol and chelation agents (Ahmed and Holmström, 2014). However, the siderophores production by halophilic microorganisms has not been studied yet in great detail (Figueroa et al. 2015; Matsui and Nishino 2016).
Halophile microorganisms have not been studied as intensively as other extremophiles although they produce halophilic enzymes and secondary metabolites that can be used for bioremediation of contaminated ecosystems, the production of polymers (Ma et al. 2010) and in the food industry (Purohit et al. 2014). It is important that enzymes used in the industry are stable when the salt and organic solvent content, pH, and temperature are high in the production process. Consequently, the isolation of enzymes from haloalkaliphilic bacteria will expand the existing industrial enzyme catalogue.
The recently dried-out maar in the volcano “Hoya Rincón de Parangueo” is part of the maar lakes of the Trans-Mexican Volcanic Belt. The sediment of the dried-out maar has a high pH, salt, aragonite and hydromagnesite content (Kienel et al. 2009), which makes it an interesting environment to search for novel extreme haloalkaliphilic microorganisms with unique metabolic capacities. The objective of this study was to isolate bacteria from this extreme haloalkaline environment that produce extracellular enzymes or siderophores with a potential industrial application.
Methods
Sampling sites and sample collection
Three sediment samples were collected from the 0-20 cm layer of a dried-out maar in the volcano “Hoya Rincón de Parangueo”, Valle de Santiago in the state of Guanajuato, Mexico (20º 43´N, 101º 25´W) (Fig. 1) on July 2015, mixed and stored at 4º C until used. The study site was characterized by high electrolytic conductivity (EC) and high alkalinity (Kienel et al. 2009). Details of the physicochemical characteristics can be found in Ibarra-Sánchez et al. (2019).
Isolation of extremophilic bacteria on artificial growth media
Halotolerant heterothophic aerobic bacteria were grown in the liquid media A, B, C, D, E, F and G (Supplementary Table 1) that contained 20% NaCl. The different media were selected to enrich and isolate bacteria with a wide variety of metabolic preferences. Sediment sub-samples (10 g) were inoculated separately in Erlenmeyer flasks containing the different liquid media (90 mL) with 20% NaCl and pH 9 and incubated at room temperature and 110 rpm for 7 days except for media H, I and J (Supplementary Table 1). After incubation, the enriched cultures were serially diluted 10-4 to 10-6 in 15% NaCl solution and 0.1 mL aliquots were plated on Petri dishes containing each of the same agar-media. The Petri dishes were incubated at room temperature for 7 to 15 days. Sediment samples were placed on agar-medium H and J in Petri dish, while soil extract was mixed with agar in medium I. Colonies were selected based on their morphology and purified on the same media after several re-inoculations. Colonies from media F, G and I were purified in medium D due to its size were larger in this medium.
Biochemical characteristics of the isolates
Bacterial strains were examined for Gram staining, catalase and oxidase. The growth of the isolates at different concentrations of NaCl was studied by growing them in a medium containing (g L-1 of distilled water) peptone 5.0, yeast extract 2.0, beef extract 1.0, agar 18.0, and varying amounts of NaCl (0.0% to 35%) at pH 9, for 48 h to 72 h. The growth of the isolates at different pH was studied in liquid media containing (g L-1 of distilled water) MgSO4.7H2O 1.0, NaCl 100.0, sodium citrate 3.0, yeast extract 10.0 and (NH4)2SO4 1.0 (Romano et al. 2005a) with pH ranging from 5.5 to 12 using the following buffers NaH2PO4/Na2HPO4 (pH 5.5-8.0), NaHCO3/Na2CO3 (pH 9.0-10), Na2HPO4/NaOH (pH 11.0-12.0) as reported by Dou et al. (2016). The production of different extracellular enzymes (caseinases, amylases, lipases, proteases, xylanases and cellulases) and siderophore production were tested. The different media contained ingredients as listed in Supplementary Table 2. Production of extracellular enzymes were determined after 24 h to 72 h and siderophore production were confirmed by a yellow halo around the colonies.
DNA extraction and PCR amplification
The DNA was extracted from pure cultures after 48 h at room temperature. The cells were harvested with 15% w/v NaCl and centrifuged at 7,700 ( g for 15 min. The pellets were dissolved in one mL buffer (0.25 M EDTA pH 8), 80 μL 10 mg mL-1 lysozyme was added and the mixture was incubated at 37 ºC for one h. One mL lysis solution (0.1M NaCl, 0.5 M Tris-HCl [pH 8], 12% sodium dodecyl sulphate) and 0.5 g sterile sand were added, and samples were vortexed for 10 min. The solution was treated with a cycle of freezing at -70 ºC for 20 min and thawing at 65 ºC for 20 min or 30 s at 6.0 m s-1 in FastPrep-24TM (Biomedicals, Solon, OH). The sample was centrifuged at 7,700 ( g at room temperature for 10 min. Proteins were removed from the supernatant with 1/5 volume EDTA (0.5 M, pH 8) and 1/10 volume potassium acetate (5 M, pH 5.5), incubated at 4º C for 20 min and centrifuged at 12,000 ( g at 4 ºC for 5 min. The supernatant was extracted twice with 500 μL chloroform-isoamyl alcohol (24:1 vol: vol). The solution was transferred to a clean tube and DNA precipitated with one volume 13% polyethylene glycol (MW, 8000) dissolved in 1.6 M NaCl. The mixture was incubated at -20 ºC overnight and centrifuged at 12,000 ( g at 4 ºC for 15 min. The DNA pellet was washed with cool 70% ethanol, dried and dissolved with 50 μL sterile deionized water and stored at -20 ºC.
The 16S rRNA genes of the genomic DNA were amplified with primers 27F and 1492R (Lane, 1991). The reaction mixture contained 2 mM MgCl2, 0.2 mM each dNTP, 10 µM of each primer, 10 ng DNA template and 0.2 U Taq DNA polymerase (Thermo Scientific, California, USA) with 1 ( reaction buffer in a total volume of 25 μL. The PCR was done with an initial denaturation at 95ºC for 10 min, 30 cycles of 45 s at 95 ºC, 45 s at 56 ºC and 60 s at 72 ºC, followed by a final 10 min extension at 72 ºC. The PCR products were purified using UltraClean® PCR Clean-Up kit (MoBio Laboratories, Carlsbad, CA, USA) as recommended by the manufacturer. Sequencing was done by Macrogen Inc. (DNA Sequencing Service, Seoul, Korea) in an ABI 3730XLs sequencer.
Sequence analysis
All the sequences were compared with 16S rRNA gene sequences available in the EZBioCloud database (http://www.ezbiocloud.net/, 17 july 2018). Pairwise and multiple sequence alignments of approximately 1300 bp were done using CLUSTAL W 2.0 (Larkin et al. 2007). A phylogenetic tree was constructed from evolutionary distances using the neighbour-joining clustering method. The best substitution model to calculate the genetic distances was determined with the Model Test implemented in MEGA X (Kumar et al. 2018). Tree topologies were determined by bootstrap analysis of 1000 replications in MEGA X package. The 16S rRNA gene sequences obtained have been submitted to the NCBI GenBank Database under accession numbers as MH727817 to MH727853.
Results
16S rRNA phylogenetic analysis
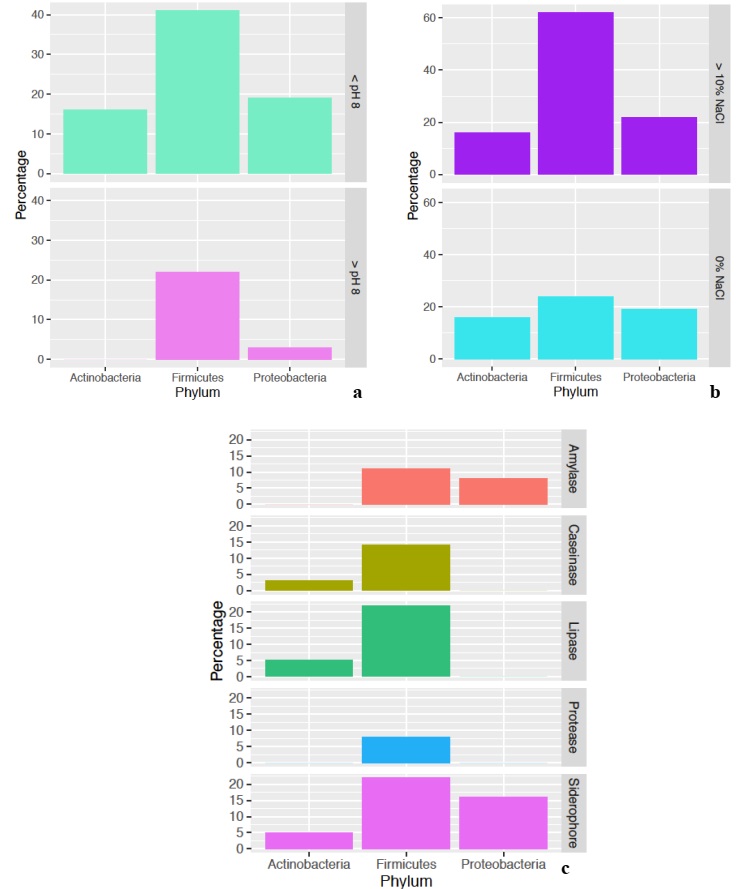
Thirty-seven strains were obtained from the dried-out maar in the volcano “Hoya Rincón de Parangueo”. Twenty-four % of the isolates grew in low nutrient medium whereas 76% were isolated in high rich medium (Supplementary Table 3). The majority of the isolates belonged to the Firmicutes (23 isolates) followed by Proteobacteria (eight isolates) and Actinobacteria (six isolates) (Fig. 2). Media A, B, C, D, E, F, I and J favoured the isolation of Firmicutes, media G and H Proteobacteria, while Actinobacteria were obtained only in the medium I (Supplementary Table 3). The isolates were classified in eight bacterial genera. The most abundant genera were Alkalibacillus (A. haloalkaliphilus, A. filiformis) (Fig. 3a) and Halomonas (H. pantelleriensis and H. fontilapidosi) (eight isolates each one) (Fig. 3b) followed by Nesterenkonia (N. lutea and N. alba) (six isolates) (Fig. 3c), Oceanobacillus (O. oncorhynchi and O. kimchii) (five isolates), Staphylococcus (S. equorum subsp. equorum) (four isolates), Bacillus (B. luteus, B. lindianensis and B. chagannorensis, B. saliphilus) (four isolates) and Salsuginibacillus (S. halophilus) (two isolates) (Fig. 3a).

Fig. 2 Bacteria isolated from the sediment of a dried-out maar in the volcano “Hoya Rincón Parangueo”, Guanajuato, Mexico at the phylum and genus level.
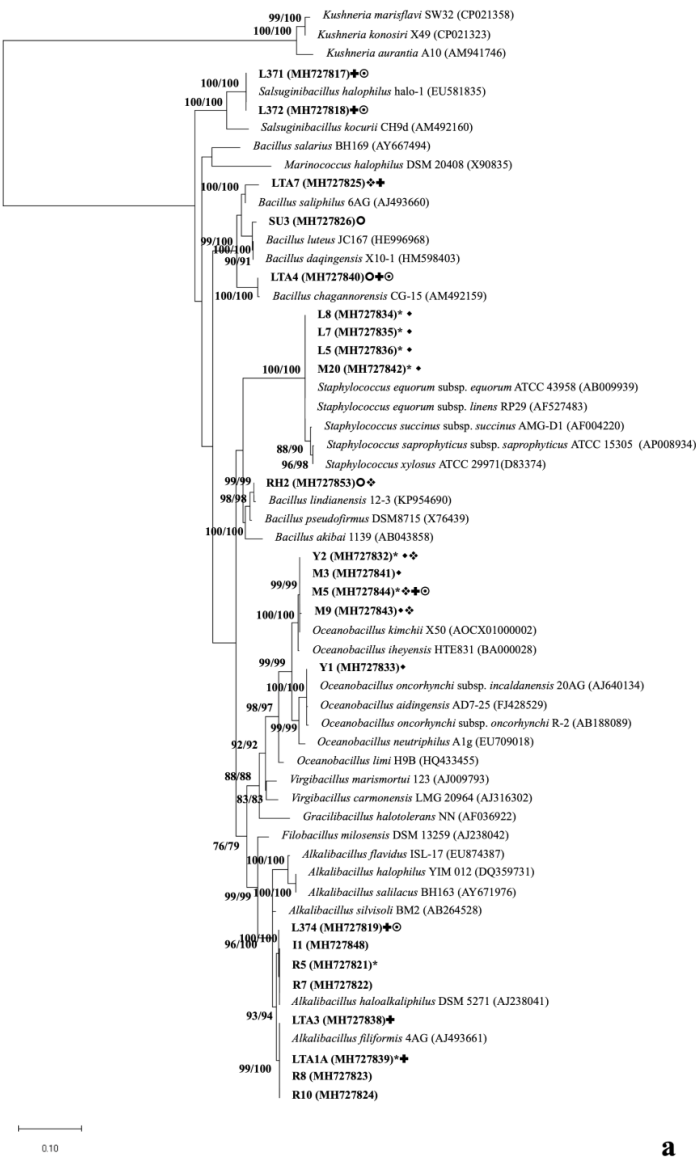
The tree was constructed with Maximum Likelihood method. The evolutionary distances were computing using 1298 bp, 1396 bp and 1300 bp using Tamura 3-parameter model for Firmicutes, Proteobacteria and Actinobacteria, respectively. The rate variation among sites was modelled with gamma distribution (shape parameter=5). All positions containing gaps and missing data were eliminated. The numbers of the branches represent percentages of bootstrap sampling of 1000 replications of neighbour-joining/maximum likelihood analysis. * Produce siderophore, ( produce lipase, ο produce amylase, ❖ produce caseinase, growth above pH 8, ʘ growth above 5% NaCl. Strains in green bold are possible new species.
Fig. 3a Phylogenetic diversity of haloalkaliphilic heterotrophic Bacteria based on comparison of 16S rRNA gene of isolates from the sediment of a dried-out maar in the volcano “Hoya Rincón Parangueo”, Guanajuato, Mexico with representatives of the EzBiocloud database. a) Members of phylum Firmicutes.
The tree was constructed with Maximum Likelihood method. The evolutionary distances were computing using 1298 bp, 1396 bp and 1300 bp using Tamura 3-parameter model for Firmicutes, Proteobacteria and Actinobacteria, respectively. The rate variation among sites was modelled with gamma distribution (shape parameter=5). All positions containing gaps and missing data were eliminated. The numbers of the branches represent percentages of bootstrap sampling of 1000 replications of neighbour-joining/maximum likelihood analysis. * Produce siderophore, ( produce lipase, ο produce amylase, ❖ produce caseinase, growth above pH 8, ʘ growth above 5% NaCl. Strains in green bold are possible new species.
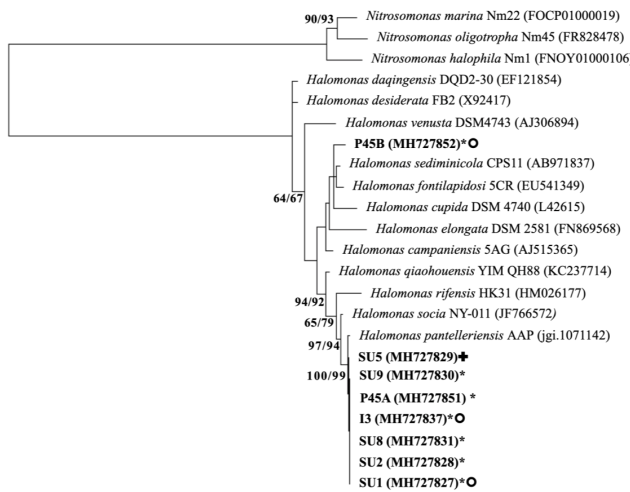
Fig. 3b Phylogenetic diversity of haloalkaliphilic heterotrophic Bacteria based on comparison of 16S rRNA gene of isolates from the sediment of a dried-out maar in the volcano “Hoya Rincón Parangueo”, Guanajuato, Mexico with representatives of the EzBiocloud database. b) Members of phylum Proteobacteria.

The tree was constructed with Maximum Likelihood method. The evolutionary distances were computing using 1298 bp, 1396 bp and 1300 bp using Tamura 3-parameter model for Firmicutes, Proteobacteria and Actinobacteria, respectively. The rate variation among sites was modelled with gamma distribution (shape parameter=5). All positions containing gaps and missing data were eliminated. The numbers of the branches represent percentages of bootstrap sampling of 1000 replications of neighbour-joining/maximum likelihood analysis. * Produce siderophore, ( produce lipase, ο produce amylase, ❖ produce caseinase, growth above pH 8, ʘ growth above 5% NaCl. Strains in green bold are possible new species.
Fig. 3c Phylogenetic diversity of haloalkaliphilic heterotrophic Bacteria based on comparison of 16S rRNA gene of isolates from the sediment of a dried-out maar in the volcano “Hoya Rincón Parangueo”, Guanajuato, Mexico with representatives of the EzBiocloud database. c) Members of phylum Actinobacteria.
Isolates RE (95.9%), RG1 (95.9%), RG2 (96.4%), RH1 (96.9%), RA (96.0%) and I11 (95.7%) belonged to Nesterenkonia and might be new species, as they had more than 3% of interspecific divergence and less than 2% intraspecific divergence (Fig. 3c, Supplementary Table 4).
Haloalkaliphilic bacteria isolation and biochemical characteristics
All isolates were catalase positive, oxidase negative except for isolates LTA7, RH2, Y1, P45A, I3, P45B and SU8, 76% were Gram-positive and 24% Gram-negative (Supplementary Table 3). All isolates grew optimally at pH 9, 24% of them could growth only at pH > 8 and 49% could growth at pH < 8 (Supplementary Table 5, Fig. 4a). All isolates grew in more than 10% NaCl, most of them grew with less than 5% NaCl and only two grew in 30% NaCl, but more than 80% could growth in 25% NaCl and 59% grew with no salt addition to the medium (Supplementary Table 5, Fig. 4b).
Haloalkaliphilic Bacteria Enzymatic activities
The bacterial isolates (37) were screened for extracellular amylases, cellulases, xylanases, caseinases, chitinases, proteases and lipases (Fig. 4c). Overall, 20 isolates (53%) produced at least one of these enzymes, but none showed activity for all of them. Amylase activity was detected in 19% of isolates belonging to A. haloalkaliphilus, B. chagannorensis, B. lindianensis, B. luteus, H. pantelleriensis (two isolates) and H. fontilapidosi. Lipase activity was found in 10 isolates belonging to S. equorum (four isolates), O. oncorhynchi, O. kimchii (three isolates) N. alba and N. lutea. Moreover, caseinase activity was detected in 16% of the isolates i.e., A. saliphilus, O. kimchii (three isolates) N. lutea and B. lindianensis (Supplementary Table 5).
Siderophore production
Siderophore production was found in 17 isolates (46%), belonging to Firmicutes (22%), Proteobacteria (19%) and Actinobacteria (5%) (Fig. 4c). They were members of S. equorum (four isolates), A. haloalkaliphilus, A. filiformis, O. kimchii (two isolates), H. pantelleriensis (six isolates), H. fontilapidosi, N. alba and N. lutea (Supplementary Table 5).
Discussion
Halotolerant microorganisms have been isolated from hypersaline environments, such as hypersaline ponds (Tang et al. 2011), saline soil (Shi et al. 2012) and different other saline environments (El Hidri et al. 2013). However, no reports exist about the isolation of haloalkaliphilic microorganisms from a dried-out maar of a volcano, such as the “Hoya Rincón de Parangueo”. In this study, isolates from Firmicutes were more abundant than those belonging to the Proteobacteria and Actinobacteria as found in other hypersaline environments, such as a saline-alkali soil from China (Tang et al. 2011; Shi et al. 2012) and a saline lake from India (Joshi et al. 2008).
The ability to produce enzymes with industrial application was tested. Lipases, which are widely used in detergent and food industry (Panda and Gowrishankar, 2005), were found only in O. oncorhynchi, O. kimchii and N. lutea. The production of lipases has been reported in halophiles, such as Staphylococcus, Oceanobacillus and Nesterenkonia (El Hidri et al. 2013; Jeong et al. 2014; Kiran et al. 2014). However, there are no reports of lipase production by species found in this study. Amylases, which are in increasing demand for their application in detergent, pharmaceutical production and wastewater treatment (Margesin and Schinner, 2001), were detected in isolates belonging to Alkalibacillus, Bacillus and Halomonas as reported previously (Romano et al. 1996; Jeon et al. 2005; Gonzalez-Domenech et al. 2009; El Hidri et al. 2013). Although caseinase activity has been reported in Bacillus, Oceanobacillus and Nesterenkonia (Romano et al. 2005b; Delgado et al. 2006; Hirota et al. 2013), there are no reports of caseinase production by O. kimchii and N. lutea as found in this study. They might be alternatives for halophilic proteases applied as laundry additives, and in pharmaceutical waste management and food processing (Razzaq et al. 2019).
Siderophores have many applications in agriculture, bioremediation techniques, microbial ecology and medicine, i.e., they are used as biocontrol agents, biosensors, to improve the mechanism of antibiotics and to improve the growth of uncultured microorganisms in the laboratory (Ahmed and Holmström, 2014; Saha et al. 2016). Siderophore production by microorganisms is well documented (Cabaj and Kosakowska, 2009; Peek et al. 2012; Lehner et al. 2013) and studies have focused mainly on Staphylococcus sp. due to its clinical importance (Dale et al. 2004; Mukherjee et al. 2017). Halophiles and alkaliphiles, such as Bacillus sp. and Halomonas sp., produce siderophores (Figueroa et al. 2015; De Serrano et al. 2016; Matsui and Nishino, 2016) and siderophores of N. jeotgali, N. massiliensis and Nesterenkonia sp. are involved in mechanisms of pathogenicity through iron acquisition (Chander et al. 2017). However, no evidence of siderophore production by N. alba, Oceanobacillus and Alkalibacillus have been reported previously.
Bacillus sp., Nesterenkonia sp., Halobacillus sp., Filobacillus sp., Micrococcus sp., Salinivibrio sp., Halomonas sp., Streptomyces sp. and Virgibacillus sp. are halophiles known to produce halophilic hydrolases with industrial applications. In this study, Alkalibacillus and Halomonas were the most abundant genera with a known biotechnological application as they produce ectoine, PHA and hydrolases (Bergmann et al. 2013; Yin et al. 2015). Additionally, some of the microorganisms isolated in this study produce siderophores and amylases with promising industrial application.
Members of Nesterenkonia was the third most abundant genus isolated from the dried-out maar in the volcano “Hoya Rincón de Parangueo”. Some strains belonging to Nesterenkonia are haloalkaliphilic found in hypersaline natural environments, industry effluents and fermented seafood (Chander et al. 2017). At the time of writing, the genus Nesterenkonia include 20 species with validated names with 14 of them isolated from natural hypersaline environments, i.e. N. halobia (Mota et al. 1997), N. lacusekhoensis (Collins et al. 2002), N. xinjiangensis, N. halotolerans (Li et al. 2004), N. lutea, N. sandarakina (Li et al. 2005), N. aethiopica (Delgado et al. 2006), N. halophila (Li et al. 2008), N. rhizosphaerae (Wang et al. 2014), N. alkaliphila (Zhang et al. 2015), N. aurantica (Finore et al. 2016), N. cremea (Sultanpuram et al. 2017), N. pannonica (Borsodi et al. 2017) and N. natronophila (Machin et al. 2019). Isolates found in this study are grouped with N. alba, a strain isolated from black liquor of a cotton pulp (Luo et al. 2009). Members of Nesterenkonia are known to produce halophilic proteases and xylanases (Yin et al. 2015), but no evidence of siderophore production has been reported as found in this study. Based on a cut-off of 97% identity for 16S rRNA gene, commonly used to discriminate bacterial species (Tindall et al. 2010), the isolates from the cultured Nesterenkonia could be new species. Further studies will be necessary to get the physiological and biochemical characteristics of these isolates to differ them from the recognized species.
Finally, isolates obtained from “Hoya Rincón de Parangueo” can grow at very different pH, temperature, and salt concentration compare with those found in other volcano craters in Mexico, such as “El Chichón” where acidophilic bacteria were isolated (Ovando-Chacón, et al. 2020) or Xinantécatl volcano where psychrophilic microorganisms are found (Tapia-Vázquez et al. 2020). However, lipase production was detected in bacteria from both “Hoya Rincón de Parangueo” and "El Chichón" despite the pH difference of each environment. Moreover, siderophore production was found in isolates from the Xinantécatl volcano, a cold environment, as well as in this study.
In conclusion, strains isolated from the dried-out maar in the volcano “Hoya Rincón de Parangueo” were haloalkaliphilic or halotolerant. Six isolates belonging to genus Nesterenkonia could be new species. Siderophore production was detected for the first time in extreme microorganisms belonging to N. alba, Alkalibacillus and Oceanobacillus.
Conflict of interest
The authors declare that they have no conflict of interest.











 nueva página del texto (beta)
nueva página del texto (beta)