Introduction
Land use by humans that results in habitat loss and fragmentation (e.g., urbanization, deforestation, agriculture) is one of the most significant drivers of biodiversity loss. Human-induced land and climate change also has a negative impact by altering patterns of climatic and abiotic variables that affect species distribution patterns (Molina-Sánchez et al., 2019; Pauls et al., 2013; Parusnath et al., 2017). Biodiversity encompasses variation at all levels, namely within and among ecosystems, communities, species, and populations. Nonetheless, most efforts regarding the monitoring and conservation of such diversity have been focused on ecosystems and species, while intraspecific variation has been usually overlooked, of which genetic variation is a fundamental component (Hoban et al., 2020; Mimura et al., 2017). Genetic variation underpins population fitness and adaptive potential and is key in terms of species extinction risk (Hoffmann et al., 2017; Reed & Frankham, 2003). Importantly, habitat loss and fragmentation decreases the size and connectivity of populations, with a consequent loss of genetic diversity at both species and population levels (Allendorf et al., 2012; DiBattista, 2008). Safeguarding genetic variation is therefore essential to mitigating biodiversity loss (Leigh et al., 2019; Pereira et al., 2013; Sarre & Georges, 2009).
Similarly, identification and mapping of habitats and of the connectivity of the landscape matrix for a given species are critical components for the conservation of populations and species (Underwood et al., 2013). In turn, the degree to which a landscape facilitates or limits movement of individuals between habitats is tightly linked with individual gene flow (Schoville et al., 2012). Accordingly, to evaluate the effects of fragmentation and environmental heterogeneity at the genetic level, 2 main information sources are needed: the landscape features (e.g., climate, topography) along the distribution of populations, at the scale of interest, and the genetic parameters of individuals (e.g., gene flow) (Allendorf et al., 2012; Balkenhol et al., 2015). Landscape genetics allows precisely that, by assessing the effects of landscape composition, configuration, and heterogeneity on microevolutionary processes, by measuring functional connectivity, and evaluating fine-scale population structure, in particular gene flow (Balkenhol et al., 2015; Schoville et al., 2012). It considers the landscape as a heterogeneous area and it is focused on the organisms’ specific environmental variables (temperature, precipitation, and so forth) and/ or physical characteristics (topography, rivers, roads) that together define landscape heterogeneity (Balkenhol et al., 2015; Garrido-Garduño & Vázquez-Domínguez, 2013).
One common problem in ecological, landscape, and conservation studies is that one often must work with little-known, endangered, or threatened species, or with insufficient a priori or in situ information regarding species’ distributions, habitat heterogeneity, and the environmental features that govern their occurrence (Balkenhol & Fortin, 2015). When the concern or interest is determining the dispersal of such species, this lack of information hinders the possibility of adequately establishing the landscape characteristics potentially affecting the movement of individuals (and gene flow). Furthermore, it significantly limits designing the appropriate sampling schemes to do it. These problems are exacerbated where, due to current conservation urgency, it is imperative to obtain rapid and yet robust data about environmental features and landscape heterogeneity.
As one way to tackle this problem, we here propose to apply an environmental domains method to identify the climatic and environmental characteristics that define a landscape matrix across the distribution of the species of interest, at different spatial scales, and where in situ information is scarce. Environmental domains, defined as discrete areas with similar environments (Leathwick, 1998; Leathwick et al., 2003), is a framework that allows the classification of environmental sites in which non- biological elements, like climatic variables, are used to explain biological patterns and generate hypotheses (Téllez-Valdés & Dávila-Aranda, 2003; Tellez-Valdés et al., 2010). For instance, environmental domains have been used to select sets of sites that represent regional species diversity, namely sites that encompass the highest number of different groups of species or communities (Londoño-Murcia et al., 2010; Téllez-Valdés et al., 2010), or with a more land-use focus to find suitable crop sites and for agrobiodiversity (Parra-Quijano et al., 2012). The novelty of our approach is, on the one hand, that it can be built on a range of geographic scales, overcoming the limitations posed by the traditional description of the climate (García, 1998), which does not allow differentiating the climate categories at multiple scales. Accordingly, it can be applied at the different scales of, among others, ecological, wildlife management, biogeographic or conservation studies. On the other, even when there is little in situ environmental information, our approach can be explicitly used at the local scale to identify localities that encompass the most heterogeneous sites (highest environmental differences among them) across that landscape for the target species.
Here, we specifically tested the suitability of our approach to define a sampling strategy for a landscape genetics study, using as focal species the rodent Heteromys pictus for which we had previously obtained genetic data. We demonstrate that such a sampling strategy can capture environmental differences of the landscape matrix potentially associated with a species’ genetic structuring (Balkenhol et al., 2015; Schoville et al., 2012), allowing us to evaluate functional connectivity under a landscape genetics framework (i.e., where individual dispersal is measured indirectly via gene flow). Heteromys pictus (formerly placed in the genus Liomys) is an endemic heteromyid species from western and southern Mexico, predominantly distributed in tropical dry forests (TDFs) along the Pacific coast. TDFs harbor significant levels of biological diversity, with nearly 19% of Mesoamerican endemic fauna inhabiting these forests (Miles et al., 2006). They are characterized by a marked seasonality with long dry periods and a short rainy season, which renders an ecosystem dominated by high temporal and spatial habitat heterogeneity, and where a variety of species with specific adaptations to climatic pressures are concentrated (Murphy & Lugo, 1986; Olson & Dinerstein, 1998). A few studies have shown the tight relationship between the landscape features and environmental heterogeneity with the ecological patterns and genetic structure of rodent species in TDFs (Garrido-Garduño et al., 2016; Vázquez-Domínguez et al., 1998, 1999; Vega et al., 2017).
Our approach, the methods and results of which we report here, followed overall these steps: we applied the environmental domains method to define heterogeneous landscape sites across the distribution of the species of study, at 3 nested geographic scales: 1) national (Mexico), to encompass the country’s widest environmental differences, 2) regional (along the Mexican Pacific coast), to identify environmental heterogeneity along the entire distribution of Heteromys pictus, and 3) local, at 2 protected tropical dry forest areas, which we targeted for future landscape genetics evaluations with H. pictus. Next, at the local scale we obtained a comprehensive set of climatic and environmental variables that defined the landscape matrix (the 2 protected areas), based on which we designed a sampling strategy and selected sampling localities for H. pictus, which were corroborated on the field. These sampling localities indeed comprised the broadest habitat heterogeneity, capturing the configuration and heterogeneity of the landscape matrix, and allowing us to identify key environmental variables significantly associated with dispersal of H. pictus individuals across their natural habitat (Garrido-Garduño et al., 2016).
Materials and methods
Environmental heterogeneity at different scales
We worked at 3 spatial scales: national, regional and local, which allowed us to consider the environmental variability of the domains at nested geographic levels (Fig. 1). To demonstrate how our approach permits the assessment of hierarchical levels of classification, we first evaluated a national scale to particularly comprise the entire country’s extraordinary heterogeneity resulting from factors like climate, lithology, orography, geological history, hydrology, and the vast diversity of vegetation types (e.g., perennial, cloud, coniferous and tropical deciduous forests, desert scrub, grassland, wetlands) (Challenger & Soberón, 2008). The regional scale included the Pacific coast, defined based on the environmental domains at the national scale and where Heteromys pictus is distributed (see below). At the local scale we chose 2 localities (natural protected areas) from within the regional Pacific coast domain. One was the Estación Biológica Chamela (EBC) on the coast of Jalisco (3,319 ha), which is characterized by low hills and shallow creeks, a marked seasonality with annual precipitation (731 mm), and a mean temperature of 24.6 °C. The dominant vegetation is tropical deciduous forest with occasional areas of semi-deciduous forest along the courses of seasonal streams (Rzedowski, 1978; Challenger & Soberón, 2008). The second locality was Parque Nacional Huatulco (PNH; 6,374 ha) in southern Mexico on the coast of Oaxaca, with deciduous forest as the dominant vegetation. PNH has a complex orography that promotes isolation and high species diversity, with warm subhumid climate, 90% rainfall in summer, and a 28 °C mean annual temperature.
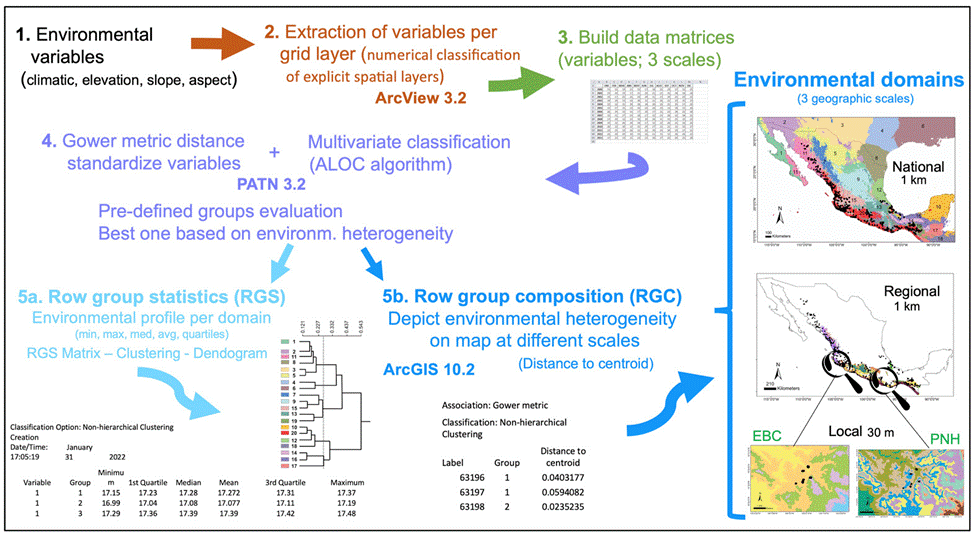
Figure 1 Diagram depicting the steps and methods followed to obtain the environmental domains for the 3 scales (global, regional, local). See the detailed explanation in the Methods section.
The environmental variables for analyses were chosen based on the following conditions: they were from freely available sources and captured the climatic and environmental variability, in order to describe the landscape heterogeneity at different scales (Fig. 1). Accordingly, the environmental layers for the national and regional scales were based on a Digital Elevation Model (DEM) GTOPO30 (at an original spatial resolution of 0.0083333 km2), obtained from the United States Geological Service (https://webgis.wr.usgs.gov/globalgis/gtopo30/gtopo30.htm); they were aggregated to a factor 3 to obtain a spatial resolution of 9 km2 for statistical analyses. We included the area along the southern United States to the north, and Guatemala, Belize, a region of El Salvador, and Honduras to the south, to encompass a continuous and consistent coverage of the climate of Mexico and to assure a confident interpolation. For the EBC and PNH areas, the DEM was obtained from the ASTER Global Digital Elevation Map Announcement (http://asterweb.jpl.nasa.gov/gdem.asp), on 30 x 30 m grid cells (90 m2 spatial resolution). We used the 19 bioclimatic variables interpolated by Cuervo-Robayo et al. (2013) for Mexico, to build environmental layers for the national and regional scales (Supplementary material Table S1), whereas for the local scale we used 17 climatic variables because the BIO14 and BIO17 had values of zero, becoming uninformative. We also included 3 layers of terrain variables (elevation, slope, and aspect) for all scales.
Classification of environmental domains
The sequential steps we performed to obtain the environmental domains are depicted in figure 1. Based on a numerical classification of explicit spatial layers (Leathwick et al., 2003), we extracted the different environmental variables per grid layer with the Grid Analyst Extension in Arcview 3.2. We built a data matrix for the national scale with 578,421 cells corresponding to the maximum, mean, and minimum values for the 19 environmental variables, while the regional scale matrix included 28,226 cells (see below for further detail), both at 9 km2 spatial resolution. However, a very different environmental data set was interpolated for the local scale, formed by 25,920 and 258,121 cells (at 90 m2) for EBC and PNH, respectively. The Gower metric distance measure was used to standardize the environmental variables, which permitted the combination of variables with different measurement units (Gower, 1971; Sneath & Sokal, 1973). Next, we performed for each scale a multivariate classification based on the standardized distance measurements, with the ALOC algorithm that builds a non-hierarchical classification (designed for large data sets), using the program PATN 3.2 (https://patn.org/f-a-q/) (Belbin, 1987, 1993, 1995). The number of groups or clusters that PATN produces is user-defined, controlled by selecting the minimum permissible distance between any pair of “seed-sites”, on the basis of a dissimilarity measure (Belbin, 1993). We evaluated different pre- defined groups and chose the one that best encompassed the corresponding environmental heterogeneity at national and regional scales, aided by known global vegetation and climate classifications (Rzedowski, 1978; García, 1998). The result is an intergroup similarity matrix with which to examine the structure of each group, producing classifications unambiguously determined by specific variables. Another advantage of this method is that it makes the classification objective across the study area replicable (Metzger et al., 2005). Two files are obtained with the latter classification: row group composition (RGC) and row group statistics (RGS). The RGS yields the environmental profile of each domain, which is the average of all the variables used for the localities at each scale (minimum, maximum, median, average, 1st and 3d quartile values; see Fig. 1), from which a matrix is built. The dissimilarity between domains was estimated from the RGS matrix with a cluster analysis and the resulting relationships are shown as a dendrogram. The RGC data allowed depicting the landscape’s environmental heterogeneity on a map, at different scales, using ArcGIS 10.2.1. Finally, we obtained species occurrences of Heteromys pictus from the Global Biodiversity Information Facility (GBIF; www.gbif.org) and plotted them on the national and regional scale maps, whereas for the local scale we used our own H. pictus sampling individuals obtained from fieldwork.
To specifically test our approach for its suitability to define a sampling strategy for a landscape genetics study, we used the rodent Heteromys pictus. Based on our initial environmental classification results with the highest dissimilarity values at the national scale, we selected the domain in which H. pictus is present as the regional species distribution frame (see Results). From the multivariate classification results at this regional scale, we selected 2 domains that each encompassed the EBC and PNH áreas for our local scale (Fig. 1). We pre-defined 5 and 10 groups for the local scale multivariate classification analysis, with the purpose of exhibiting the high heterogeneity existing at even this small scale. To evaluate the performance (i.e., accuracy for depicting the highest heterogeneity) of the environmental domains classification, we followed 3 procedures: first, we went to the field to both EBC and PNH domains and confirmed overall differences among them as the general floristic composition (O. Téllez-Valdés performed the identification), vegetation cover (%), elevation (m asl), and the landscape matrix in between (rivers, primary and secondary roads). Secondly, we extracted the bioclimatic profile from latitude and longitude points and, derived from the environmental variables that defined each domain, we performed a principal components analysis (PCA; Borcard et al., 2011), using the function “princomp” in the stats library v.3.2.0 in R (R Core Team, 2015). The PCA was done to reduce redundant environmental variables and obtain those that best explained the environmental variation between domains. With the results of this environmental profile we used the randomization and permutation procedure analysis of similarities (ANOSIM) developed by Clarke (1993) to statistically evaluate differences between domains, and to estimate the between- and the within- group mean rank similarities (R). The method uses the Bray-Curtis measure of similarity, and the null hypothesis is that there are no differences between the members of the various domains. To test for significance, the ranked similarity within and between domains was compared with the similarity that would be generated by random chance. Essentially, the samples are randomly assigned to groups 1,000 times and R is calculated for each permutation. The observed R value is then compared against the random distribution to determine if it is significantly different from that which could occur at random (Clarke, 1993). As a result, ANOSIM displays the degree of separation between groups and generates a value of R, where R = 0 suggest no differentiation and R = 1 indicates complete differentiation (Clarke & Warwick, 2001). ANOSIM was run using the function “anosim” in the vegan library v 2.2-1 (Oksanen, 2015) in R.
Next, we used the 5 and 10 domains obtained for EBC and their environmental profiles to design the sampling scheme -selection of sampling localities encompassing the broadest heterogeneity- for the landscape genetics study with Heteromys pictus. Namely, we estimated the landscape matrix differences among the selected sampling localities, and with genetic data we had previously obtained for H. pictus (published in Garrido-Garduño et al., 2016), we evaluated the gene flow between individuals across the landscape matrix. Briefly, we evaluated how environmental and landscape features shaped the genetic structure at a fine scale among H. pictus populations from each sampling site (104 individuals, 6 sampling localities,14 microsatellite loci), by assessing the effect of the landscape variables on patterns of gene flow. A detailed description of the entire genetic protocol, genetic analyses, and landscape statistics is provided in Garrido-Garduño et al. (2016).
Results
The chosen area at the national scale encompassed a total of 20 environmental domains, including southern USA and northern Central America, where the dendrogram shows 2 broad groups, Central-North (CN) and South Pacific-Gulf (SPG), with a between dissimilarity value of 0.5426 (Fig. 2a, b). The CN included 3 internal groups, while the SPG had 2. Within SPG, domain 20 (see numbers assigned to each domain in Fig. 2b), named Coastal Pacific, encompassed the geographic distribution of Heteromys pictus. In accordance, the Coastal Pacific domain was chosen as our next, regional scale. At this scale, environmental heterogeneity was captured as 2 groups with 6 domains (Fig. 2c, d), which coincided with deciduous forest vegetation along the Pacific coast where H. pictus is distributed, including the 2 protected areas of our local scale (domains 5 and 6). Notably, it also encompassed an isolated region of semi-deciduous forest on the Gulf of Mexico (domain 2), which is the limit of the eastern distribution of H. pictus.
The domains classification results for each EBC and PNH (Fig. 3; Supplementary material Tables S2-S5) captured the wide environmental heterogeneity at the local scale; as an example, we show the variation in precipitation exhibited at both the 5 and 10 domains (Fig. 4). Furthermore, the PCA results supported the existence of a robust environmental difference among the selected domains (Fig. 5) within each EBC and PNH. The first 2 axes of the PCA comprised 98.9% of the environmental variation in EBC, exhibiting a strong, and consistent, separation between domains for the 5 (Fig. 5a) and 10 (Fig. 5b) domains classification. The environmental variables that contributed the most to the differentiation were precipitation of the wettest quarter and mean temperature of wettest quarter for the first axis, and temperature seasonality for the second axis. Results were similar for PNH, where the first 2 axes of the PCA comprised 96.7% of the environmental variation, showing a consistent and clear separation between domains for the 5 and 10 domains classification (Fig. 5c, d). Precipitation of wettest quarter and minimum temperature of the coldest period, for the first axis, and precipitation seasonality for second axis, were the environmental variables that contributed the most.
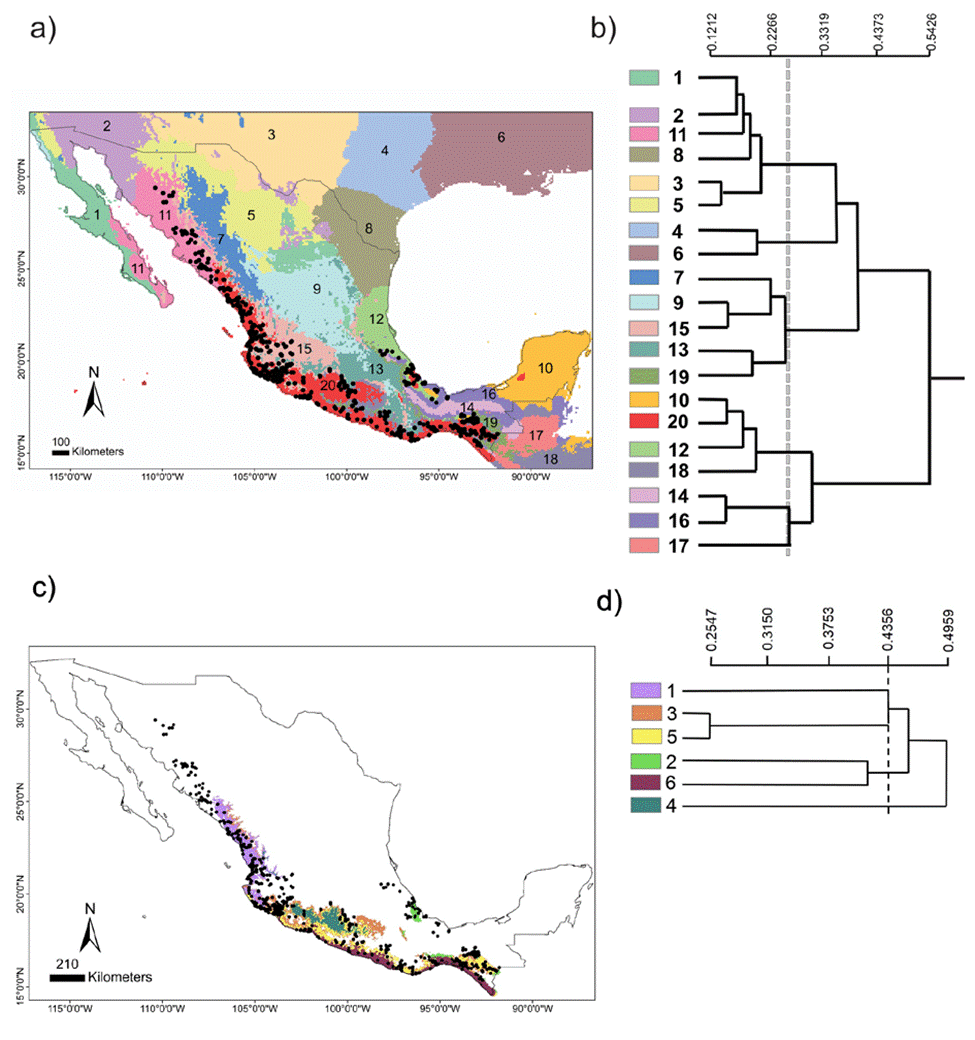
Figure 2 National scale: (a) geographical distribution of the 20 environmental domains for Mexico, numbered 1 to 20 and depicted by different colors. Black circles represent the location of species occurrences taken from the Global Biodiversity Information Facility (GBIF; www.gbif.org) for Heteromys pictus. Regional scale: (c) geographical distribution of the 6 environmental domains (from the Coastal Pacific, the environmental domain associated with the distribution of Heteromys pictus; see Results), numbered 1 to 6 and depicted by different colors (colors in the online version). Location of the Estación Biológica Chamela (EBC) and the Parque Nacional Huatulco (PNH) areas is shown. (b, d) Dendrogram depicting similarity levels between domains (similarity values shown on the upper scale).
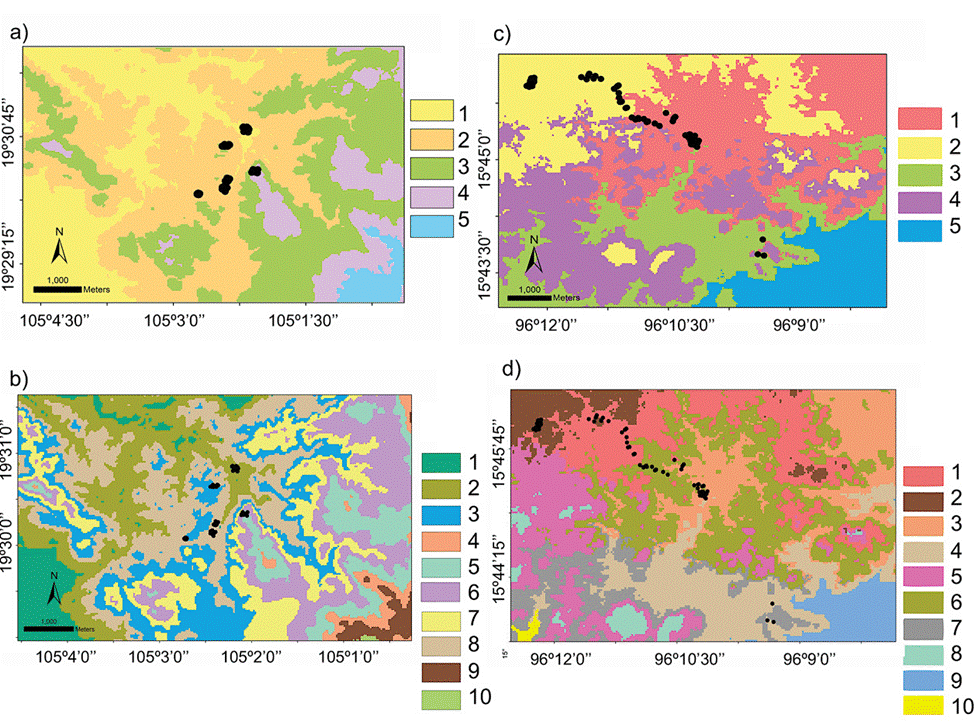
Figure 3 Geographical distribution of the environmental domains at the local scale for the Estación Biológica Chamela (EBC) and the Parque Nacional Huatulco (PNH), of the 5 (a, c; numbered 1 to 5) and 10 classification (b, d; numbered 1 to 10) respectively, depicted by different shades of grey (colors in the online version). Black circles represent sampling localities for Heteromys pictus individuals from our fieldwork (Garrido-Garduño et al., 2016).
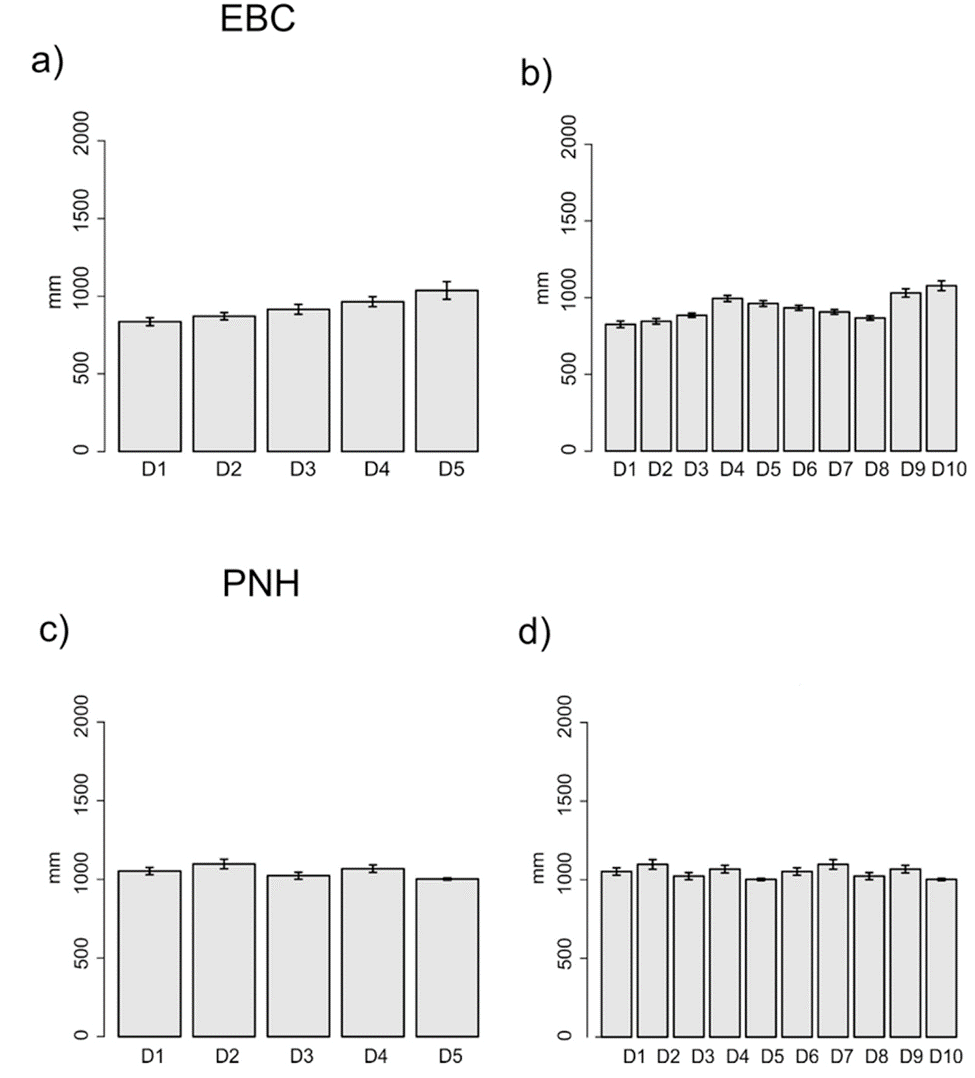
Figure 4 Annual mean precipitation of each sampling locality for the 5 (a, c) and 10 (b, d) environmental domains classification, for the Estación Biológica Chamela (EBC) and Parque Nacional Huatulco (PNH), respectively.
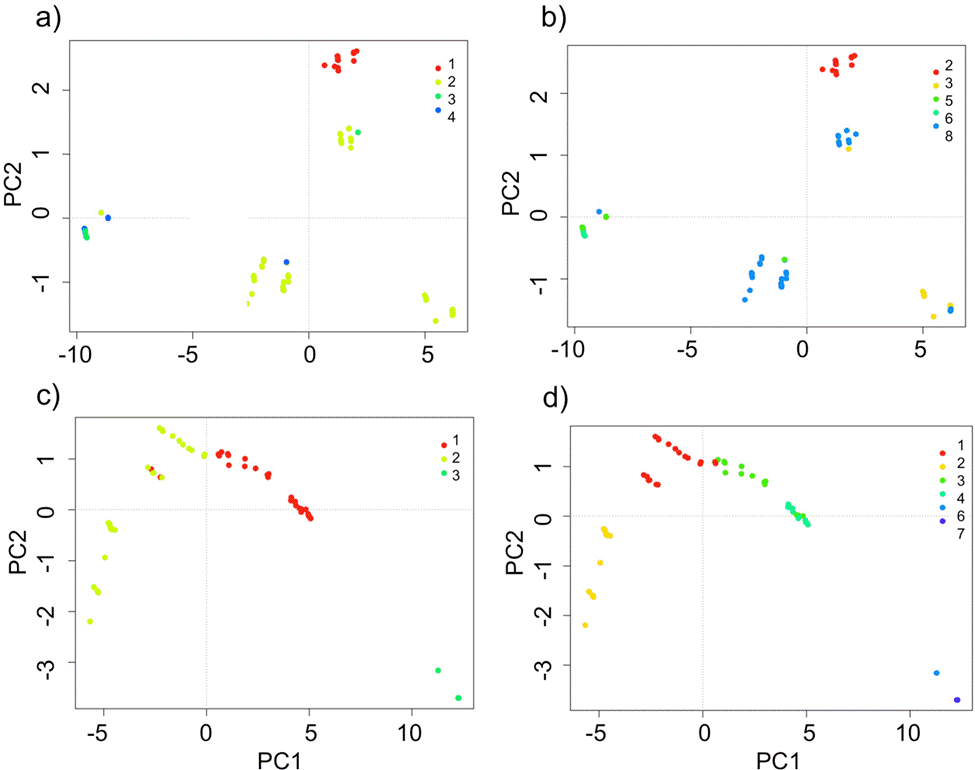
Figure 5 Principal component analysis (PCA) for the 5 and 10 environmental domains classification, at the local scale, for the Estación Biológica Chamela (EBC) (a, b) and Parque Nacional Huatulco (PNH) (c, d). Different domains indicated by their numbers in the insert (top right corner) and depicted by different shades of grey (colors in the online version).
The ANOSIM results for both EBC and PNH confirmed that the environmental composition differed among the 5 domains classification (R = 0.412, p < 0.001; R = 0.830, p < 0.001) (Table 1), thus were significantly different from random (Clarke, 1993). The pairwise analysis showed, for EBC, that the highest difference was between domains 1 and 4 (R = 0.984) and 1 and 3 (R = 0.962) (p < 0.001), while the lowest was between domains 4 and 3 (R = 0.039; p = 0.362). PNH showed strong differences between all comparisons among domains 1, 2, and 3 (p < 0.001). Moreover, the ANOSIM for the 10 domains classification in EBC (Table 2) showed a medium degree of separation at the global level (R = 0.554; p < 0.001) and high for some pairwise comparisons, namely between domains 6 and 2, and 6 and 3, which exhibited the highest difference recorded (R = 1; p < 0.001); the lowest was between domains 5 and 6 (R = 0.198; p < 0.037). Regarding PNH for the 10 domains, the ANOSIM results showed a higher degree of separation at the global level (R = 0.858; p < 0.001) and between all domain comparisons (values from 1 to 0.628; p < 0.001).
Table 1 ANOSIM results for the selected environmental domains for the 5 domains classification at the local scale for (a) Estación Biológica Chamela (EBC) and (b) Parque Nacional Huatulco (PNH). Some domains are not shown due to lack of data for comparison.
| a) | EBC | |||||
| Domain 1 (D1) | Domain 2 (D2) | Domain 3 (D3) | ||||
| R | p | R | p | R | p | |
| D1 | ||||||
| D2 | 0.0717 | 0.110 | ||||
| D3 | 0.9623 | 0.001 | 0.6403 | 0.001 | ||
| D4 | 0.9845 | 0.001 | 0.7478 | 0.001 | 0.0397 | 0.362 |
| Global | 0.412 | 0.001 | ||||
| b) | PNH | |||||
| Domain 1 (D1) | Domain 2 (D1) | |||||
| R | p | R | p | |||
| D1 | ||||||
| D2 | 0.8091 | 0.001 | ||||
| D3 | 0.9633 | 0.001 | 0.999 | 0.001 | ||
| Globa | 0.8309 | 0.001 | ||||
Table 2 ANOSIM results for the selected environmental domains for the 10 domains classification at the local scale for (a) Estación Biológica Chamela (EBC) and (b) Parque Nacional Huatulco (PNH). Some domains are not shown due to lack of data for comparison.
| a) | EBC | |||||||||
| D2 | D3 | D5 | D6 | |||||||
| R | p | R | p | R | p | R | p | |||
| D2 | ||||||||||
| D3 | 0.7647 | 0.001 | ||||||||
| D5 | 0.9892 | 0.001 | 0.9662 | 0.001 | ||||||
| D6 | 1 | 0.001 | 1 | 0.001 | 0.1981 | 0.037 | ||||
| D8 | 0.2401 | 0.003 | 0.4575 | 0.001 | 0.6766 | 0.001 | 0.8714 | 0.001 | ||
| Global | 0.5548 | 0.001 | ||||||||
| b) | PNH | |||||||||
| D1 | D2 | D3 | D4 | D6 | ||||||
| R | p | R | p | R | p | R | p | R | p | |
| D1 | ||||||||||
| D2 | 0.9071 | 0.001 | ||||||||
| D3 | 0.6278 | 0.001 | 1 | 0.001 | ||||||
| D4 | 0.9985 | 0.001 | 1 | 0.001 | 0.283 | 0.003 | ||||
| D6 | 1 | 0.050 | 1 | 0.030 | 1 | 0.004 | 1 | 0.050 | ||
| D7 | 1 | 0.030 | 1 | 0.004 | 1 | 0.007 | 1 | 0.003 | 1 | 0.330 |
| Global | 0.858 | 0.001 |
Hence, based on these results, we were able to design a field sampling strategy where we selected sampling sites, at both EBC and PNH, based on the domains that had the combined greatest climatic and environmental dissimilarity between them. Importantly, we subsequently corroborated in the field that the chosen sampling localities indeed encompassed the broadest environmental heterogeneity: 6 at EBC (Fig. 3a, b; see Fig. S1 in Garrido-Garduño et al., 2016) and 3 at PNH (Fig. 3c, d).
Discussion
The importance of preserving connectivity both in protected areas and in human-altered landscapes for conservation of wild plant and animal populations is amply recognized (Foresta et al., 2016; Parusnath et al., 2017). Furthermore, Ducci et al. (2019) identified different strategies for preserving biodiversity, among which we emphasize the need to establish the ecological requirements of species and to map the distribution of their habitat, in order to identify the elements that contribute to their dispersal and connectivity.
Our environmental domains approach rendered a convenient, objective, and reproducible strategy to identify the range of environmental variation in an area, breaking the continuous change composition into discrete and complementary groups, namely differentiated domains at different spatial scales. Notably, it was based on general, freely available climatic and environmental information, not requiring detailed data acquired on site through intensive fieldwork, proving one of our aims, i.e., when in situ environmental information is scarce. Our approach is innovative in various ways, in which we can identify contrasting domains that encompass the highest heterogeneity at nested geographic levels. The latter is crucial when there is little previous environmental information, needed to design, among others, an appropriate sampling scheme for ecological studies. Moreover, from the domains obtained one can not only select contrasting sampling localities but also a comprehensive set of climatic and environmental variables that define both the landscape homogeneity and heterogeneity within and between sites, respectively. Finally, the geographic domains and environmental variables obtained characterize the features of the landscape matrix potentially affecting the connectivity and dispersal patterns of a species, and ultimately its genetic structure. Hence, those variables can be incorporated into a landscape genetics study, as we exemplified with Heteromys pictus, whilst also relevant for research at different scales (e.g., ecological and biogeographic studies, wildlife management, and conservation assessments).
Nested environmental domains capture the extent of landscape heterogeneity
The classification of our environmental domains reflects changes across landscapes, variations that can be significantly different depending on scale (Leathwick et al., 2003). In accordance, it enables identifying landscape sites that can be easily differentiated, even when some can be close to each other along the geographical or environmental space (Austin & Smith, 1989; Belbin, 1993). Indeed, we analyzed the complex environment at different scales, where the domains we obtained efficiently captured the extraordinary heterogeneity encompassed by both climate and vegetation at the country level, incorporating the intrinsic ecosystem diversity (Challenger & Soberón, 2008). Hence, we believe our approach at this scale would be a helpful tool for evaluating differences in regional studies, like biogeographic assessments or genetic structure patterns in the context of biogeographic and phylogeographic studies for species with wide distributions (e.g., Hidalgo-Galiana et al., 2014; Melville et al., 2016).
Habitat selection and adaptation to local environmental conditions may be the primary processes structuring diversity among landscapes (Perktaş et al., 2016; Quiroga et al., 2019), ultimately reflected in the genetic structure of populations. Hence, having a means to indirectly acquire information about habitat heterogeneity is crucial to enable a better understanding of correlations between, for instance, climatic variables and biological or genetic data (Morin & Lechowicz, 2008). Landscape genetics and genomics have made it possible to evaluate such correlations (Manel et al., 2003), where sampling design is a paramount aspect (Hall & Beissinger, 2014). Notably, our results show that the different number of domains at each scale analyzed exhibited diverse and specific values of climatic variables (e.g., temperature, precipitation). Landguth et al. (2011)) generated an individual-based program to simulate genetic differentiation in a spatially continuous population inhabiting a landscape with gradual resistance values to movement (dispersal), to obtain the best sampling sites across the species distribution. They simulated a wide range of combinations by varying genetic diversity measures and number of individuals sampled from the population, finding that all influenced statistical power. However, they did not incorporate into their analyses the environmental heterogeneity, which can directly influence an individual’s movement, gene flow, or adaptive genetic diversity patterns (Schoville et al., 2012). Therefore, and since the environmental domains we obtained performed significantly better than a random selection of sites (ANOSIM analyses), we prove that our approach can serve as a tool to efficiently determine a sampling design that encompasses the greatest environmental heterogeneity and the variables (quantitative data) that define it, in explored and unexplored landscapes.
Our environmental domains approach put into practice: a case study
To assure the adequacy of the environmental domains at the local scale, and because each domain can be visualized by mapping them into geographic space, we corroborated our results both in the field and by PCA and ANOSIM analyses. Hence, we had high confidence in the sampling localities chosen, which can be used for a variety of ecological, conservation, or genetic studies. We specifically aimed to prove the applicability for landscape genetics studies, where both environmental heterogeneity and landscape matrix defining variables are required (Balkenhol & Fortin, 2015).
The rodent Heteromys pictus has specialized morphological, ecological, and behavioral characteristics associated with its ability to survive in markedly seasonal environments. Although it shares the habitat with several other rodent species, it is the dominant species in the dry forest and a key element in the ecosystem, responsible for the dispersal of seeds and the recruitment dynamics of many species of plants (Vázquez-Domínguez et al., 1998, 1999). The 6 sampling sites selected based on our local domain classifications at EBC encompassed the highest heterogeneity within the local area (Figs. 3, S1 in Garrido- Garduño et al., 2016), being also of biological significance for H. pictus (Vázquez-Domínguez et al., 2002). Our results in Garrido-Garduño et al. (2016) proved that habitat heterogeneity influences H. pictus dispersal patterns and fine-scale population processes of gene flow. Specifically, we found differences in the structural and functional connectivity among sampling sites (domains), including variables like precipitation that are significantly associated with H. pictus genetic structure, movement of individuals, and gene flow, further corroborating the adequacy of our approach to define contrasting and heterogeneous sites
We also applied our environmental domains results for PNH, where we found a clear association between the landscape heterogeneity and environmental variables (precipitation, elevation) with the genetic patterns of H. pictus populations. Notably, although PNH is a protected area there is almost no biological data for it, hence our approach served as a novel tool for planning a landscape genetics sampling design with little in situ information. Recent studies have proved that gene flow (i.e., connectivity among populations) is key for the genetic rescue of small populations, essential for the conservation of biodiversity (Frankham, 2015; Mimura et al., 2016). The use of our approach could certainly span much more, providing for example a first approximation to evaluate species distribution limits, habitat suitability, and habitat associations for wildlife management and conservation. Additionally, it could be used in the reintroduction or translocation of species, where out of the many aspects that need to be considered, targeting places that are climatically similar to the species’ current distribution (Pearce & Lindenmayer, 1998) and careful knowledge of the genetics of individuals (Norman & Christidis, 2021; Shaney et al., 2020), are key aspects to minimize the risk of failure of such efforts. Hence, given the potential to identify not only heterogeneous but also climatically similar sites, a practical application in conservation biology would be to combine our environmental domains approach, ecological patterns, and landscape genetics information to design species reintroduction and translocation programs.











 nueva página del texto (beta)
nueva página del texto (beta)


