Introduction
Troglobitic organisms are those restricted to hypogean (i.e., subterranean) habitats throughout their life cycle, and as a result of this particular ecology and distribution they tend to evolve an exceptional phenotype characterized by the reduction -or even complete loss- of body pigmentation and ocular structures (Romero & Paulson, 2001; Wilkens & Strecker, 2017). The troglobitic phenotype is therefore considered by many an adaptation to cave life, as well as a classic example of regressive and convergent evolution (Porter & Crandall, 2003). Evidence from developmental biology and molecular evolution studies, however, suggests that both selection and genetic drift have played a role in the evolution of troglomorphism (Rétaux & Casane, 2013). Mexico is not only a megadiverse country in a broad sense, but it also contains one of the most diverse subterranean faunas of the world (Reddell, 1981), which includes troglobitic species of both invertebrates and vertebrates. In Mexico, invertebrate troglobites comprise species from a number of arthropod groups such as crustaceans, insects, arachnids, diplopods and chilopods, while vertebrate troglobites are taxonomically restricted to teleost fishes (Nicholas, 1962). Although troglomorphism has evolved multiple times in the tree of life, fishes are one of the groups of organisms with the largest number of taxa and lineages independently adapted to cave life (Helfman et al., 2009; Proudlove, 2006). Of the 200+ troglobitic species of fish worldwide, 10 are found in Mexico (Proudlove, 2006), 9 of which are endemic, and 1 of which, Astyanax mexicanus (or A. jordani, depending on the author), is arguably the most popular model in studies of evolutionary cave adaptation mechanisms (Gross, 2012; Miller, 2005). Notably, Mexican-endemic cave fishes comprise representatives from phylogenetically distant lineages, and while the majority can be considered primary freshwater fishes, they also include descendants from brackish/marine ancestors, such as the eleotrid Caecieleotris morrisi (Gobiiformes: Eleotridae) and the viviparous brotula Typhlias pearsei (Ophidiiformes: Dinematichthydae) (Møller et al., 2016; Walsh & Chakrabarty, 2016). In contrast to A. mexicanus, very little is known about most of Mexico’s troglobitic ichthyofauna, even on fundamental aspects such as their taxonomy and basic ecology.
Notably, 4 of the 10 troglobitic fish species present in Mexico are members of the genus Rhamdia Bleeker 1858 (Siluriformes: Heptapteridae), a catfish widely distributed in the Neotropical region, from Mexico to Argentina (Hernández et al., 2015; Perdices et al., 2002). Rhamdia is a taxonomically complex group that lacks synapomorphies and is therefore of questionable monophyly (Bockmann, 1998; DoNascimiento et al., 2004; Garavello & Shibatta, 2016). In the latest revision of the genus, Silfvergrip (1996) consolidated its diversity to only 11 species (of the 100+ considered as valid at the time), arguing that, by increasing the geographical coverage of comparative material, phenotypic discontinuities that initially allowed to differentiate between putative species faded into a spectrum of intraspecific morphological variation. Subsequent authors, however, in view of patterns of morphological, ecological, and genetic variation, did not adopt several of Silfvergrip’s nomenclatural proposals (Hernández et al., 2015; Perdices et al., 2002; Weber et al., 2003; Weber & Wilkens, 1998). Currently, Rhamdia comprises 27 valid species (Fricke et al., 2020), 6 of which are troglobitic and endemic to their respective country of occurrence: the Brazilian Rhamdia enfurnada (Bichuette & Trajano, 2005), the Venezuelan Rhamdia guasarensis (DoNascimiento et al., 2004), and the Mexican Rhamdia reddelli (Miller, 1984), Rhamdia zongolicensis (Wilkens, 1993), Rhamdia macuspanensis (Weber & Wilkens, 1998), and Rhamdia laluchensis (Weber et al., 2003). Four of the 7 species of Rhamdia present in Mexico are in fact troglobitic, with Rhamdia laticauda, Rhamdia guatemalensis, and Rhamdia parryi being the only epigean, or surface forms (Miller, 2005). Mexican troglobitic Rhamdia species occur in karstic systems in the southern portion of the country, each being restricted to its own type-locality cave (Fig. 1). Although troglobitic forms of Rhamdia have been known to exist in Mexico since the 1930s (Hubbs, 1936, 1938), the discovery and description of the 4 currently recognized cave-dwelling species ultimately resulted from specimens collected throughout the 1970s and 1980s by American and Italian explorers during speleological - not biological- surveys (Mosier, 1984; Robertson, 1983). Despite reports of troglobitic populations from other caves (Mosier, 1984), these were never investigated from a taxonomic perspective. Various authors believe that the troglobitic forms of Rhamdia in Mexico derive from the epigean species R. laticauda (Greenfield et al., 1982; Perdices et al., 2002; Silfvergrip, 1996; Wilkens, 1993), which includes a hypogean population in Belize described under the category of subspecies (R. laticauda typhla) (Greenfield et al., 1982). The evidence in support for the hypothesis that R. laticauda populations gave rise to the troglobitic Rhamdia species in Mexico, however, is presently scarce and insufficient.
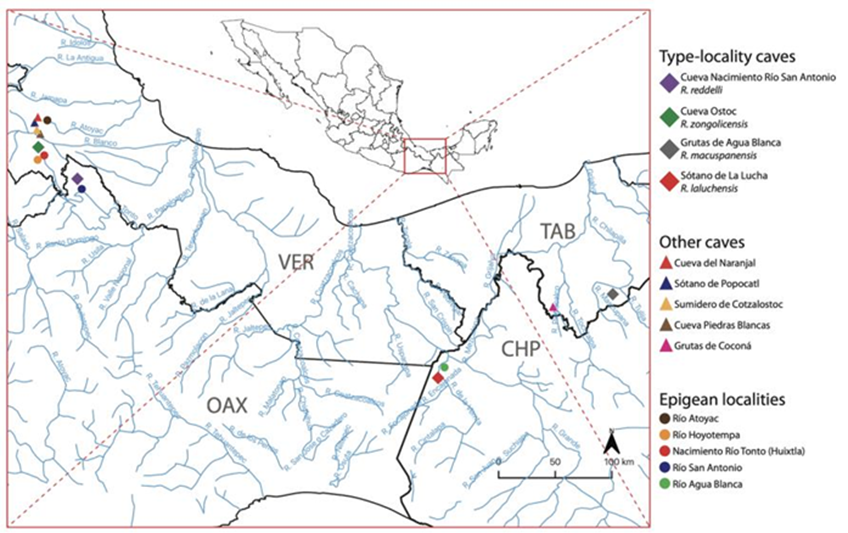
Figure 1 Map of the study area (southern Mexico) indicating the location of type-locality caves, non-type-locality caves, and epigean localities sampled during the field component of this study. Reference map based on a 2016 water bodies dataset (scale 1:50,000) from the Instituto Nacional de Estadística y Geografía (INEGI) and plotted using the geographic information system software QGIS (QGIS Development Team, 2020).
The taxonomic history of Rhamdia as it relates to its Mexican troglobitic species has not been altogether straightforward, as evidenced by a series of past synonymies and the ensuing taxonomic ambiguities. Silfvergrip (1996) was the first to cast doubt on the validity of the troglobitic species described at the time, and synonymized R. zongolicensis and R. reddelli with R. laticauda arguing a lack of diagnostic characters beyond the typical reductions/ losses associated with troglomorphism. Subsequently, in the only study to date that has investigated phylogenetic relationships and species boundaries in Middle American Rhamdia, Perdices et al. (2002) concluded that R. reddelli -the only troglobitic species included in their analysis- ought to be synonymized with R. laticauda in order to maintain a taxonomy congruent with phylogeny. More recently, Miller (2005) regarded R. zongolicensis a junior synonym of R. reddelli, while acknowledging the validity of R. macuspanensis and R. laluchensis. Interestingly, in his description of R. zongolicensis, Wilkens himself admitted that morphological differences between R. zongolicensis and the previously described R. reddelli were “minute because of convergent evolution” (Wilkens, 1993, p. 375).
It is thus self-evident that one of the main problems with the existing taxonomy of Rhamdia lies in the insufficiency of the evidence offered to justify the recognition of troglobitic species. Although the presence of morphological discontinuities is traditionally used in species descriptions as indirect evidence of divergence and reproductive isolation, the main morphological discontinuities offered for the differentiation between epigean and hypogean forms of Rhamdia are precisely those related to the regressive troglobitic phenotype, which has resulted in a lack of consensus on the exact diversity of the group and the abovementioned taxonomic ambiguities. That the presence of characters related to cave life is sufficient evidence for the distinction of species is questionable (Miller, 2005), even more so when there appears to be significant intrapopulation variation in those characters, manifested in the coexistence of normal (i.e., eyed and pigmented), troglomorphic, and phenotypically intermediate individuals (Mosier, 1984; Sbordoni et al., 1986; Wilkens, 2001). Indeed, the same type of clinal variation has been documented in A. mexicanus, and none of its troglobitic populations is considered a distinct species (Keene et al., 2015; Miller, 2005). Furthermore, current taxonomic designations do not consider molecular evidence in a phylogeographic/phylogenetic context, which for more than a decade has become a standard methodological approach of integrative taxonomic studies (Dayrat, 2005; Padial et al., 2010).
Given the manifest need for testing the adequacy of its current taxonomy, this study investigates comprehensively, for the first time, patterns of genetic and phenotypic variation in Mexican troglobitic Rhamdia in a phylogenetic context. We leverage novel collections of comparative material, including fresh specimens and tissue samples from all 4 valid cave species and other cave and surface populations/species, to shed light on the hypothesis that troglobitic forms derive from populations of the epigean species R. laticauda that opportunistically colonized underground environments in different regions, but never completely diverged/speciated from ancestral epigean populations.
Materials and methods
Voucher specimens and associated tissue samples were obtained through dedicated collecting trips to the type-locality caves of the 4 troglobitic species in southern Mexico. Whenever possible, epigean habitats -such as rivers and streams- adjacent to or in the vicinity of the type-locality caves were sampled for surface Rhamdia populations/species (Fig. 1). Hypogean specimens were primarily caught by means of minnow traps and dip nets, whereas surface forms were collected using electrofishing, cast nets, and hooks and lines. Some of the sampled cave systems were considerably deep (>50 m and up to 200 m) and not easily accessible. In those cases, sampling was accomplished with the assistance of professional speleologists well trained in vertical caving techniques (Minton & Droms, 2019). Fishes were collected and euthanized prior to preservation in accordance with recommended guidelines for the use of fishes in research (Nickum et al., 2004) and under collecting permits SGPA/ DGVS/04259/17 and SGPA/DGVS/05375/19 issued by the Mexican Ministry of Environment and Natural Resources (Secretaría de Medio Ambiente y Recursos Naturales; Semarnat). Stress and suffering were minimized by limiting handling prior to euthanasia, which was accomplished by means of the anesthetic tricaine mesylate (MS-222). Tissue samples (fin clips) were taken in the field, immediately preserved in 95% ethanol, and later cryopreserved at -80 °C. Voucher specimens were fixed in a 10% solution of formalin and subsequently transferred to 70% ethanol for long-term storage. All newly collected specimens were curated, cataloged, and deposited in the Colección Nacional de Peces (CNPE) of the Universidad Nacional Autónoma de México (UNAM). Overall, we collected a total of 129 Rhamdia specimens, including representatives of the 4 Mexican troglobitic species (R. reddelli: n = 8; R. zongolicensis: n = 9; R. macuspanensis: n = 5; R. laluchensis: n = 27), of 4 undescribed hypogean populations from the Sierra de Zongolica, Veracruz, herein treated as R. cf. zongolicensis (n = 43), and of the primarily epigean R. laticauda (n = 22) and R. guatemalensis (n = 15) (Table 1).
Table 1 List of Rhamdia specimens (and associated data) collected during the course of this study and included in genetic and morphological analyses. Mexican states abbreviated using ISO 3166 codes.
| Taxon | Habitat | Catalog | Voucher(s) | n | Locality/site ID | Lat | Long | Municipality | State | Drainage |
| R. reddelli | Cave | CNPE-IBUNAM 23807 | JA854-861 | 8 | Cueva Nacimiento Río San Antonio | 18.48 | -96.64 | Cañada San Antonio | OAX | Papaloapan |
| R. zongolicensis | Cave | CNPE-IBUNAM 23800 | JA785-791 | 7 | Cueva Ostoc | 18.61 | -96.89 | Zongolica | VER | Papaloapan |
| CNPE-IBUNAM 23806 | JA838-839 | 2 | Cueva Ostoc | 18.61 | -96.89 | Zongolica | VER | Papaloapan | ||
| R. cf. zongolicensis | Cave | CNPE-IBUNAM 23799 | JA763-784 | 22 | Sumidero de Cotzalostoc | 18.70 | -96.92 | Totolacatla | VER | Papaloapan |
| CNPE-IBUNAM 23802 | JA794-801 | 8 | Sótano de Popocatl | 18.76 | -96.95 | Coetzala | VER | Papaloapan | ||
| CNPE-IBUNAM 23817 | DADF2018.1-2018.7 | 7 | Sótano de Popocatl | 18.76 | -96.95 | Coetzala | VER | Papaloapan | ||
| CNPE-IBUNAM 23816 | DADF2015 | 1 | Sótano de Popocatl | 18.76 | -96.95 | Coetzala | VER | Papaloapan | ||
| CNPE-IBUNAM 23801 | JA792-793 | 2 | Cueva Piedras Blancas | 18.67 | -96.90 | Zongolica | VER | Papaloapan | ||
| CNPE-IBUNAM 23803 | JA802-804 | 3 | Cueva del Naranjal | 18.78 | -96.96 | Naranjal | VER | Papaloapan | ||
| R. macuspanensis | Cave | CNPE-IBUNAM uncat. | JA742 | 1 | Grutas de Agua Blanca | 17.62 | -92.47 | Macuspana | TAB | Usumacinta |
| CNPE-IBUNAM 23814 | JA932-935 | 4 | Grutas de Agua Blanca | 17.62 | -92.47 | Macuspana | TAB | Usumacinta | ||
| R. laluchensis | Cave | CNPE-IBUNAM uncat. | JA743-747 | 5 | Sótano de La Lucha | 17.06 | -93.90 | La Lucha | CHP | Grijalva |
| CNPE-IBUNAM 23811 | JA898-919 | 22 | Sótano de La Lucha | 17.06 | -93.90 | La Lucha | CHP | Grijalva | ||
| R. laticauda | Surface | CNPE-IBUNAM 23804 | JA805-809 | 5 | Río Atoyac | 18.91 | -96.78 | Atoyac | VER | Papaloapan |
| CNPE-IBUNAM 23805 | JA831-834 | 4 | Río Hoyotempa | 18.60 | -96.89 | Cuahuixtlahuac | VER | Papaloapan | ||
| CNPE-IBUNAM 23810 | JA886-896 | 11 | Nacimiento Río Tonto (Huixtla) | 18.59 | -96.88 | Aticpac | VER | Papaloapan | ||
| CNPE-IBUNAM 23809 | JA864 | 1 | Río San Antonio | 18.46 | -96.63 | Cañada San Antonio | OAX | Papaloapan | ||
| CNPE-IBUNAM 23813 | JA928 | 1 | Río Agua Blanca | 17.07 | -93.87 | La Lucha | CHP | Grijalva | ||
| R. guatemalensis | Surface | CNPE-IBUNAM 23808 | JA862-863 | 2 | Río San Antonio | 18.46 | -96.63 | Cañada San Antonio | OAX | Papaloapan |
| CNPE-IBUNAM 23812 | JA897, 920-927, 929 | 10 | Río Agua Blanca | 17.07 | -93.87 | La Lucha | CHP | Grijalva | ||
| Cave | CNPE-IBUNAM 23815 | JA943-945 | 3 | Grutas de Coconá | 17.56 | -92.93 | Teapa | TAB | Grijalva |
Novel comparative DNA sequence data -mtDNA markers cytochrome c oxidase subunit I (COI) and cytochrome b (CYB)- were generated from 27 samples, as follows: R. reddelli (n = 2), R. zongolicensis (n = 2), R. cf. zongolicensis (n = 6), R. macuspanensis (n = 2), R. laluchensis (n = 2), R. laticauda (n = 8), and R. guatemalensis (n = 8). Total genomic DNA was extracted from fresh tissue samples using the Qiagen DNeasy Tissue Extraction Kit following the manufacturer’s protocol. Amplification and sequencing of COI and CYB were carried out using the primer pairs LCO1490/HCO2198 (Folmer et al., 1994) and CytbL14841/CytbH15915 (Irwin et al., 1991; Kocher et al., 1989), respectively. DNA sequencing was carried out at Laboratorio de Secuenciación Genómica de la Biodiversidad y de la Salud (Instituto de Biología, UNAM), in-house Sanger sequencing facilities. Contig assemblage and sequence editing was performed using Geneious Prime 2020.0.4 (https://www.geneious.com). In addition to the genetic data generated from specimens collected in the field during the course of this study, we mined available COI and CYB data from Rhamdia generated in previous studies from public repositories such as The Barcode of Life Data System (BOLD) (Ratnasingham & Hebert, 2007) and GenBank. Overall, we generated/mined COI and CYB sequence data for a total of 109 specimens from 14 Rhamdia species, including all 7 distributed in Mexico. GenBank/BOLD accession numbers for both novel and preexisting sequences and their corresponding voucher specimen data are referenced in Table 2.
Table 2 Rhamdia samples (catalog and voucher) used for genetic analyses and their corresponding GenBank/BOLD accession numbers. Acronyms of ichthyological collections follow Sabaj (2020). Superscripts following accession numbers correspond to the sequence source as follows: 1) this study, 2) Arroyave et al. (2020), 3) Bermingham et al. (Unpublished), 4) BOLD (Unpublished), 5) de Carvalho et al. (2011), 6) Usso et al. (2019), 7) García-Dávila et al. (2015), 8) Hernández et al. (2015), 9) Perdices et al. (2002), 10) Pereira et al. (2013), 11) Ríos et al. (2017), and 12) Valdez-Moreno et al. (2009). Countries and provinces/states abbreviated using ISO 3166 codes.
| Species | Catalog | Voucher | Country | State | Drainage | Versant | Lat | Long | COI | CYB |
| Rhamdia reddelli | CNPE-IBUNAM 23807 | JA854 | MX | OAX | Papaloapan | A | 18.48 | -96.64 | MW4760781 | MW4675851 |
| Rhamdia reddelli | CNPE-IBUNAM 23807 | JA856 | MX | OAX | Papaloapan | A | 18.48 | -96.64 | - | MW4675861 |
| Rhamdia reddelli | MNCN 2781 | MNCN-2781 | MX | OAX | Papaloapan | A | 18.48 | -96.64 | - | AY0366979 |
| Rhamdia | CNPE-IBUNAM 23800 | JA786 | MX | VER | Papaloapan | A | 18.61 | -96.89 | MW4760791 | MW4675871 |
| zongolicensis | ||||||||||
| Rhamdia | CNPE-IBUNAM 23800 | JA787 | MX | VER | Papaloapan | A | 18.61 | -96.89 | - | MW4675881 |
| zongolicensis | ||||||||||
| Rhamdia cf. | CNPE-IBUNAM 23799 | JA764 | MX | VER | Papaloapan | A | 18.70 | -96.92 | MW4760751 | MW4675761 |
| zongolicensis | ||||||||||
| Rhamdia cf. | CNPE-IBUNAM 23799 | JA765 | MX | VER | Papaloapan | A | 18.70 | -96.92 | - | MW4675771 |
| zongolicensis | ||||||||||
| Rhamdia cf. | CNPE-IBUNAM 23801 | JA792 | MX | VER | Papaloapan | A | 18.67 | -96.90 | - | MW4675791 |
| zongolicensis | ||||||||||
| Rhamdia cf. | CNPE-IBUNAM 23801 | JA793 | MX | VER | Papaloapan | A | 18.67 | -96.90 | - | MW4675801 |
| zongolicensis | ||||||||||
| Rhamdia cf. | CNPE-IBUNAM 23802 | JA794 | MX | VER | Papaloapan | A | 18.76 | -96.95 | - | MW4675811 |
| zongolicensis | ||||||||||
| Rhamdia cf. | CNPE-IBUNAM 23803 | JA802 | MX | VER | Papaloapan | A | 18.78 | -96.96 | - | MW4675781 |
| zongolicensis | ||||||||||
| Rhamdia | CNPE-IBUNAM 23814 | JA932 | MX | TAB | Usumacinta | A | 17.62 | -92.47 | - | MW4675821 |
| macuspanensis | ||||||||||
| Rhamdia | CNPE-IBUNAM 23814 | JA934 | MX | TAB | Usumacinta | A | 17.62 | -92.47 | MW4760761 | MW4675831 |
| macuspanensis | ||||||||||
| Rhamdia | CNPE-IBUNAM 23814 | JA935 | MX | TAB | Usumacinta | A | 17.62 | -92.47 | MW4760771 | MW4675841 |
| macuspanensis | ||||||||||
| Rhamdia | CNPE-IBUNAM 23811 | JA900 | MX | CHP | Grijalva | A | 17.06 | -93.90 | MW4760671 | MW4675701 |
| laluchensis | ||||||||||
| Rhamdia | CNPE-IBUNAM 23811 | JA901 | MX | CHP | Grijalva | A | 17.06 | -93.90 | - | MW4675711 |
| laluchensis | ||||||||||
| Rhamdia | CNPE-IBUNAM 22978 | A05 | MX | CHP | Lacantún | A | 16.10 | -91.01 | MW4760691 | - |
| laticauda | ||||||||||
| Rhamdia | CNPE-IBUNAM 22978 | F04 | MX | CHP | Lacantún | A | 16.10 | -91.01 | MW4760701 | - |
| laticauda | ||||||||||
| Rhamdia | CNPE-IBUNAM 22978 | G04 | MX | CHP | Lacantún | A | 16.10 | -91.01 | MW4760711 | - |
| laticauda | ||||||||||
| Rhamdia | CNPE-IBUNAM 22978 | H04 | MX | CHP | Lacantún | A | 16.10 | -91.01 | MW4760721 | - |
| laticauda | ||||||||||
| Rhamdia | CNPE-IBUNAM 23011 | D04 | MX | CHP | Lacantún | A | 16.11 | -90.94 | MW4760731 | MW4675731 |
| laticauda | ||||||||||
| Rhamdia | CNPE-IBUNAM 23804 | JA805 | MX | VER | Papaloapan | A | 18.91 | -96.78 | - | MW4675741 |
| laticauda | ||||||||||
| Rhamdia | CNPE-IBUNAM 23805 | JA831 | MX | VER | Papaloapan | A | 18.60 | -96.89 | MW4760681 | MW4675721 |
| laticauda | ||||||||||
| Rhamdia | STRI 11528 | STRI-11528 | PA | 1 | Róbalo | A | 9.07 | -82.29 | MG9371773 | - |
| laticauda | ||||||||||
| Rhamdia | CNPE-IBUNAM 23810 | JA887 | MX | VER | Papaloapan | A | 18.59 | -96.88 | MW4760741 | MW4675751 |
| laticauda | ||||||||||
| Rhamdia | STRI 16400 | STRI-16400 | PA | 2 | Antón | P | 8.60 | -80.14 | MG9371803 | - |
| laticauda | ||||||||||
| Rhamdia | MNCN 10266-GU | MNCN10266-GU | GT | CQ | Lempa | P | 14.53 | -89.42 | - | AY0367179 |
| laticauda | ||||||||||
| Rhamdia | MNCN 3499-MEX | MNCN3499-MEX | MX | VER | Catemaco | A | 18.62 | -96.87 | - | AY0366969 |
| laticauda | ||||||||||
| Rhamdia | STRI 2686 | STRI-2686 | PA | 2 | Indio | A | 8.65 | -80.12 | MG9371753 | - |
| laticauda | ||||||||||
| Rhamdia | MNCN 46-IST | MNCN46-IST | PA | 4 | Chiriqui | P | 8.42 | -82.68 | - | AY0367289 |
| laticauda | Viejo | |||||||||
| Rhamdia | STRI 6873 | STRI-6873 | PA | 1 | Róbalo | A | 9.04 | -82.29 | MG9371743 | - |
| laticauda | ||||||||||
| Rhamdia | MNCN 653-MEX | MNCN653-MEX | MX | OAX | Papaloapan | A | 17.68 | -97.09 | - | AY0366989 |
| laticauda | ||||||||||
| Rhamdia | MNCN GU-3 | MNCNGU-3 | MX | CHP | Usumascinta | A | 16.10 | -91.77 | - | AY0367049 |
| laticauda | ||||||||||
| Rhamdia | STRI 10084 | STRI-10084 | PA | 2 | Cocle del | A | 8.70 | -80.45 | MG9371783 | AY0367329 |
| laticauda | Norte | |||||||||
| Rhamdia | STRI 11527 | STRI-11527 | PA | 1 | Róbalo | A | 9.07 | -82.29 | MG9371763 | AY0367319 |
| laticauda | ||||||||||
| Rhamdia | STRI 13659 | STRI-13659 | NI | AS | Escondido | A | 12.01 | -84.67 | MG4962173 | AY0367119 |
| laticauda | ||||||||||
| Rhamdia | STRI 13805 | STRI-13805 | NI | CO | San Juan | A | 12.00 | -85.17 | - | AY0367139 |
| laticauda | ||||||||||
| Rhamdia | STRI 14536 | STRI-14536 | NI | SJ | San Juan | A | 11.52 | -84.83 | - | AY0367129 |
| laticauda | ||||||||||
| Rhamdia | STRI 2161 | STRI-2161 | CR | A | San Juan | A | 10.91 | -85.21 | MG4962143 | AY0367149 |
| laticauda | ||||||||||
| Rhamdia | STRI 2162 | STRI-2162 | CR | A | San Juan | A | 10.91 | -85.21 | - | AY0367159 |
| laticauda | ||||||||||
| Rhamdia | STRI 218 | STRI-218 | CR | L | Sixaloa | A | 9.60 | -82.80 | MG4962133 | AY0367279 |
| laticauda | ||||||||||
| Rhamdia | STRI 219 | STRI-219 | CR | L | Sixaloa | A | 9.60 | -82.80 | - | AY0367169 |
| laticauda | ||||||||||
| Rhamdia | STRI 2686 | STRI-2686 | PA | 2 | Indio | A | 8.65 | -80.12 | - | AY0367339 |
| laticauda | ||||||||||
| Rhamdia | STRI 6345 | STRI-6345 | PA | 4 | Chiriqui | P | 8.67 | -82.85 | - | AY0367309 |
| laticauda | Viejo | |||||||||
| Rhamdia | STRI 662 | STRI-662 | PA | 4 | Chiriqui | P | 8.76 | -82.83 | MG9371813 | AY0367299 |
| laticauda | Viejo | |||||||||
| Rhamdia | STRI 722 | STRI-722 | PA | 2 | Antón | P | 8.42 | -80.25 | MG9371793 | AY0367349 |
| laticauda | ||||||||||
| Rhamdia | STRI 7821 | STRI-7821 | GT | AV | Usumacinta | A | 15.83 | -90.33 | - | AY0367059 |
| laticauda | ||||||||||
| Rhamdia | STRI 7822 | STRI-7822 | GT | AV | Usumacinta | A | 15.83 | -90.33 | - | AY0367069 |
| laticauda | ||||||||||
| Rhamdia | STRI 8099 | STRI-8099 | GT | PE | Belize | A | 16.96 | -89.36 | - | AY0367079 |
| laticauda | ||||||||||
| Rhamdia | STRI 8100 | STRI-8100 | GT | PE | Belize | A | 16.96 | -89.36 | - | AY0367089 |
| laticauda | ||||||||||
| Rhamdia | STRI 8199 | STRI-8199 | GT | IZ | Polochic | A | 15.54 | -88.90 | - | AY0367099 |
| laticauda | ||||||||||
| Rhamdia | STRI 8284 | STRI-8284 | GT | AV | Polochic | A | 15.32 | -89.86 | - | AY0367109 |
| laticauda | ||||||||||
| Rhamdia | STRI 8345 | STRI-8345 | HN | CP | Ulua | A | 14.65 | -88.88 | - | AY0367219 |
| laticauda | ||||||||||
| Rhamdia | STRI 8393 | STRI-8393 | HN | CP | Copán | A | 14.86 | -89.12 | - | AY0367209 |
| laticauda | ||||||||||
| Rhamdia | STRI 8585 | STRI-8585 | HN | YO | Aguan | A | 15.48 | -86.67 | - | AY0367229 |
| laticauda | ||||||||||
| Rhamdia | STRI 8729 | STRI-8729 | HN | OL | Patuca | A | 14.40 | -85.93 | - | AY0367249 |
| laticauda | ||||||||||
| Rhamdia | STRI 8794 | STRI-8794 | HN | OL | Patuca | A | 14.58 | -86.29 | - | AY0367239 |
| laticauda | ||||||||||
| Rhamdia | STRI 8965 | STRI-8965 | HN | FM | Choluteca | P | 14.01 | -87.00 | - | AY0367259 |
| laticauda | ||||||||||
| Rhamdia | STRI 8966 | STRI-8966 | HN | FM | Choluteca | P | 14.01 | -87.00 | - | AY0367269 |
| laticauda | ||||||||||
| Rhamdia | STRI 2122 | STRI-2122 | CR | G | Higuerón | P | 10.34 | -85.08 | MG4962153 | AY0367189 |
| nicaraguensis | ||||||||||
| Rhamdia | STRI 2121 | STRI-2121 | CR | G | Higuerón | P | 10.34 | -85.08 | MG4962163 | AY0367199 |
| nicaraguensis | ||||||||||
| Rhamdia parryi | ECOSC7185 | ECOSC7185 | MX | OAX | Tehuantepec | P | 15.41 | -92.83 | FSCH070-144 | - |
| Rhamdia parryi | ECOSC7194 | ECOSC7194 | MX | OAX | Tehuantepec | P | 16.50 | -95.06 | FSCH072-144 | AY0366999 |
| Rhamdia parryi | ECOSC7261 | ECOSC7261 | MX | OAX | Tehuantepec | P | 16.50 | -94.44 | FSCH071-144 | - |
| Rhamdia parryi | MNCNGU 178 | MNCNGU-178 | GT | ES | Acomé | P | 14.32 | -90.98 | - | AY0367029 |
| Rhamdia parryi | STRI 7723 | STRI-7723 | GT | SU | Nahualate | P | 14.50 | -91.41 | - | AY0367009 |
| Rhamdia parryi | STRI 7724 | STRI-7724 | GT | SU | Nahualate | P | 14.50 | -91.41 | - | AY0367019 |
| Rhamdia parryi | STRI 7776 | STRI-7776 | GT | ES | María Linda | P | 14.20 | -90.71 | - | AY0367039 |
| Rhamdia | IMCN-TE00018 | IMCN-TE00018 | CO | - | San Juan | P | - | - | - | KM4890748 |
| saijaensis | ||||||||||
| Rhamdia | IMCN-TE00019 | IMCN-TE00019 | CO | - | San Juan | P | - | - | - | KM4890758 |
| saijaensis | ||||||||||
| Rhamdia | IMCN-TE00020 | IMCN-TE00020 | CO | - | San Juan | P | - | - | - | KM4890768 |
| saijaensis | ||||||||||
| Rhamdia | IMCN-TE00021 | IMCN-TE00021 | CO | - | San Juan | P | - | - | - | KM4890778 |
| saijaensis | ||||||||||
| Rhamdia | IMCN-TE00022 | IMCN-TE00022 | CO | - | San Juan | P | - | - | - | KM4890788 |
| saijaensis | ||||||||||
| Rhamdia | STRI 12103 | STRI-12103 | EC | - | Daule | P | - | - | - | AY0367359 |
| cinerascens | ||||||||||
| Rhamdia | STRI 12104 | STRI-12104 | EC | - | Daule | P | - | - | - | AY0367369 |
| cinerascens | ||||||||||
| Rhamdia | CIUA 8494 | CIUA-8494 | CO | CAL | Cauca | P | 5.06 | -75.71 | UDEA184-184 | - |
| guatemalensis | ||||||||||
| Rhamdia | CIUA 8669 | CIUA-8669 | CO | ANT | Atrato | P | 6.40 | -76.71 | UDEA187-184 | - |
| guatemalensis | ||||||||||
| Rhamdia | CNPE-IBUNAM 23233 | JA012 | MX | YUC | YP aquifer | A | 20.58 | -89.61 | MW0025601 | MW0104461 |
| guatemalensis | ||||||||||
| Rhamdia | CNPE-IBUNAM 23316 | JA323 | MX | ROO | YP aquifer | A | 20.28 | -87.48 | MW4760621 | MW4675651 |
| guatemalensis | ||||||||||
| Rhamdia | CNPE-IBUNAM 23808 | JA862 | MX | OAX | Papaloapan | A | 18.46 | -96.63 | MW4760631 | MW4675661 |
| guatemalensis | ||||||||||
| Rhamdia | CNPE-IBUNAM 23812 | JA897 | MX | CHP | Grijalva | A | 17.07 | -93.87 | MW4760641 | MW4675671 |
| guatemalensis | ||||||||||
| Rhamdia | CNPE-IBUNAM 23812 | JA920 | MX | CHP | Grijalva | A | 17.07 | -93.87 | MW4760651 | MW4675681 |
| guatemalensis | ||||||||||
| Rhamdia | CNPE-IBUNAM 23815 | JA943 | MX | TAB | Grijalva | A | 17.56 | -92.93 | MW4760661 | MW4675691 |
| guatemalensis | ||||||||||
| Rhamdia | ECOCH 5785 | MX528 | GT | AV | Cahabón | A | 15.78 | -90.34 | EU75195912 | - |
| guatemalensis | ||||||||||
| Rhamdia | ECOCH 7593 | MXV-771 | BZ | 2 | - | A | 17.28 | -88.66 | MXV775-154 | - |
| guatemalensis | ||||||||||
| Rhamdia | STRI 1207 | STRI-1207 | CR | P | Cañas | P | 10.35 | -85.17 | MG4962183 | AY0366719 |
| guatemalensis | ||||||||||
| Rhamdia | STRI 13519 | STRI-13519 | NI | CI | Estero Real | P | 12.95 | -86.85 | MG4962203 | - |
| guatemalensis | ||||||||||
| Rhamdia | STRI 1669 | STRI-1669 | PA | KY | Mandinga | A | 9.47 | -79.09 | MG9371883 | AY0366859 |
| guatemalensis | ||||||||||
| Rhamdia | STRI 2165 | STRI-2165 | CR | A | San Juan | A | 10.91 | -85.21 | MG4962213 | AY0366609 |
| guatemalensis | ||||||||||
| Rhamdia | STRI 1569 | STRI-1569 | CO | CHO | Atrato | A | 5.62 | -76.74 | - | AY0366899 |
| guatemalensis | ||||||||||
| Rhamdia | STRI 816 | STRI-816 | CO | BOL | Magdalena | A | 9.36 | -74.72 | - | AY0366929 |
| guatemalensis | ||||||||||
| Rhamdia | STRI 7831 | STRI-7831 | GT | AV | Usumacinta | A | 15.83 | -90.33 | - | AY0366289 |
| guatemalensis | ||||||||||
| Rhamdia | STRI VZ-1 | STRI-VZ-1 | VE | V | Lake | A | 8.84 | -71.98 | - | AY0366949 |
| guatemalensis | Maracaibo | |||||||||
| Rhamdia laukidi | IMCN-TE00032 | IMCN-TE00032 | CO | MET | Meta- | A | - | - | - | KM4890818 |
| Orinoco | ||||||||||
| Rhamdia laukidi | IMCN-TE00034 | IMCN-TE00034 | CO | MET | Meta- | A | - | - | - | KM4890838 |
| Orinoco | ||||||||||
| Rhamdia quelen | MZUEL 14359-4477 | MZUEL14359-4477 | AR | E | Lower | A | - | - | KU8456876 | - |
| Parana | ||||||||||
| Rhamdia quelen | LBP 29359 | LBP29359 | BR | SP | Upper | A | -22.64 | -44.62 | GU70211910 | - |
| Parana | ||||||||||
| Rhamdia quelen | MCP 44783 | MCP44783 | BR | MG | São | A | -19.47 | -43.87 | HM9060115 | - |
| Francisco | ||||||||||
| Rhamdia quelen | IIAP 470 | IIAP-470 | PE | - | Amazon | A | - | - | KT9524557 | - |
| Rhamdia quelen | ROM uncat. | TZGAA016-06 | GY | BA | Waini | A | 7.50 | -59.55 | TZGAA016-064 | - |
| Rhamdia quelen | STRI 517 | STRI-517 | PE | MDD | Amazon | A | -11.80 | -71.44 | - | AY0367419 |
| Rhamdia quelen | - | VZ-54 | VE | - | Orinoco | A | 8.25 | -69.55 | - | AY0367379 |
| Rhamdia quelen | STRI 2308 | STRI-2308 | AR | W | Paraná | A | -28.53 | -58.91 | - | AY0367429 |
| Rhamdia quelen | - | P1881 | UY | RO | - | A | 39.32 | -53.93 | - | KP79869911 |
| Rhamdia voulezi | UEL 16503 | BRH013-16 | BR | PR | Iguaçu | A | -25.62 | -52.62 | BRH013-164 | - |
| Rhamdia voulezi | UEL 16503 | BRH014-16 | BR | PR | Iguaçu | A | -25.62 | -52.62 | BRH014-164 | - |
| Rhamdia voulezi | UEL 16503 | BRH015-16 | BR | PR | Iguaçu | A | -25.62 | -52.62 | BRH015-164 | - |
| Rhamdia | UEL 16503 | BRH001-16 | BR | PR | Iguaçu | A | -25.62 | -52.62 | BRH001-164 | - |
| branneri | ||||||||||
| Rhamdia | UEL 16504 | BRH002-16 | BR | PR | Iguaçu | A | -25.62 | -52.62 | BRH002-164 | - |
| branneri |
The monophyletic status, phylogenetic placement, and relationships of Mexican troglobitic Rhamdia species -with respect to each other and to epigean congeners throughout the geographic range of the genus- was investigated via phylogenetic analysis. Specifically, we used 3 different datasets based on the abovementioned comparative DNA sequence data -a COI matrix (645 bp × 53 ingroup terminals), a CYB matrix (1110 bp × 84 ingroup terminals), and a COI +CYB concatenated matrix (1755 bp × 28 ingroup terminals)- to infer phylogenetic relationships in Rhamdia, emphasizing representation of Middle American diversity and Mexican troglobitic forms (Table 2). DNA sequence data from each gene/ marker were independently aligned via multiple sequence alignment using the software MUSCLE (Edgar, 2004) under default parameters. The concatenated matrix was taxonomically limited to terminals (voucher specimens) for which both COI and CYB data were available, assembled by linking together individual alignments using the software 2matrix (Salinas & Little, 2014). Best-fit models of nucleotide substitution were statistically selected using the software jModelTest2 (Darriba et al., 2012) based on the Akaike Information Criterion and the following settings for the computation of likelihood scores: number of substitution schemes = 3, base frequencies = +F, rate variation = +I and +G (with nCat = 4), base tree for likelihood calculations = ML optimized, and base tree search = best. Individual gene alignments (i.e., COI and CYB datasets) were each analyzed partitioned by codon position, while the concatenated alignment was analyzed partitioned by both gene and codon position. Maximum likelihood (ML) inference of phylogeny for each dataset was carried out with the software RAxML-HPC BlackBox (v. 8.2.12) (Stamatakis, 2006) as implemented through the CIPRES Science Gateway (v. 3.3) (http://www.phylo.org). Nodal support was assessed using bootstrap percentage values calculated via RAxML’s rapid bootstrapping heuristics (Stamatakis et al., 2008). All trees were rooted at Pimelodella chagresi (COI: MG937103; CYB : AY036748). To further investigate species boundaries in Middle American Rhamdia -with emphasis on troglobitic forms- we analyzed comparative COI sequence data in a phenetic context (i.e., overall genetic similarity) under a DNA barcoding threshold-based approach (Hebert et al., 2003, 2004; Ward, 2009). To this end, we first computed a matrix of Kimura 2-parameter (K2P) genetic distances from the abovementioned COI matrix using the R (www.r-project.org) package “ape” (Paradis et al., 2004; Popescu et al., 2012). We used K2P-corrected genetic distances to allow broader comparisons as this metric represents the standard in DNA barcoding literature (Čandek & Kuntner, 2015). The resulting corrected distance matrix was used as input to generate a dendrogram using the hierarchical clustering algorithm Unweighted Pair Group Method with Arithmetic mean (UPGMA) (Sokal & Michener, 1958) as implemented in the R package phangorn (Schliep, 2011).
Variation in the degree of troglomorphism (i.e., body depigmentation and eye reduction) in hypogean populations was documented and assessed qualitatively by imaging external morphology dorsally, based on a sample of 4 ethanol-preserved specimens per locality, representing the spectrum of variation at each hypogean locality. Localities consisted of all 4 type-locality caves plus 2 caves from the Sierra de Zongolica, Veracruz, harboring undescribed troglobitic populations (i.e., Sótano de Popocatl and Sumidero de Cotzalostoc). Variation in external morphology and overall body shape among cave-dwelling species (including R. cf. zongolicensis from Sótano de Popocatl and Sumidero de Cotzalostoc) and available closely related epigean species (R. laticauda and R. guatemalensis) was investigated quantitatively under a multifarious approach comprising meristics, traditional morphometrics (TM), and landmark-based geometric morphometrics (GM). Variation in overall body shape was assessed on both lateral and dorsal profiles. For meristic and morphometric analyses, novel collections were complemented with 6 voucher specimens of troglobitic species from historical collections, namely R. reddelli (CNPE-IBUNAM 2440; n = 3), R. macuspanensis (CNPE-IBUNAM 22287; n = 1), and R. laluchensis (CNPE-IBUNAM 18455; n = 2). On the other hand, undescribed hypogean populations from “El Naranjal” and “Piedras Blancas” caves (Sierra de Zongolica, Veracruz) were not considered for morphological analyses, due to their small sample sizes (n = 3 and n = 2, respectively).
A total of 6 meristic and 38 mensural (linear) traits were recorded from a sample of 122 specimens: 116 of the 129 newly collected plus the abovementioned 6 from historical collections (Table 3; Fig. 2). Eight of the 129 newly collected specimens were unavailable for meristics and TM analyses because they were cryopreserved (-80 °C) immediately after photographic documentation and tissue sampling, and are being kept ultrafrozen for future genomic studies. Selection of meristic and linear traits was informed by relevant previous taxonomic work (Hernández et al., 2015; Silfvergrip, 1996). Linear traits were measured with a handheld digital caliper to the nearest 0.01 mm, standardized as a proportion of either standard length (SL) or head length (HL), and log-transformed for use in TM analyses. Exploratory analysis of linear data was conducted via Principal Component Analysis (PCA), seeking to visualize general patterns of morphological variation among species (including 2 undescribed cave-dwelling populations provisionally identified as R. cf. zongolicensis) and to identify morphometric variables that contribute to the distinction of groups within the multivariate space. To test for significant differences in overall body shape among species/populations, a Multivariate Analysis of Variance (MANOVA) was conducted using the resulting first 3 principal components (PCs) as dependent variables, and species/populations as independent (predictor) variables. Lastly, post hoc pairwise comparisons (Bonferroni corrected) were conducted as follow-up tests of differentiation (Hotelling’s T 2 test) between pairs of species/populations. Both PCA and MANOVA analyses (including post hoc comparisons) were carried out with the software PAST (v. 4.04) (Hammer et al., 2001).
Table 3 Meristic and linear morphometric variables used to assess external morphological variation.
| Variable type | Variable | Abbreviation |
| Linear | Standard length | SL |
| Head depth | HD | |
| Body depth at dorsal fin | BD | |
| Snout to posterior tip of occipital process | S-OP | |
| Head length | HL | |
| Snout to insertion of dorsal fin | S-DF | |
| Snout to insertion of adipose fin | S-AdF | |
| Snout to insertion of pectoral fin | S-PcF | |
| Snout to insertion of pelvic fin | S-PvF | |
| Snout to anal cavity | S-A | |
| Snout to insertion of anal fin | S-AF | |
| Pectoral fin spine length | PcFSL | |
| Dorsal fin spine length | DFSL | |
| Mouth length | ML | |
| Inter-maxillary barbel distance | IMBD | |
| Head width | HW | |
| Body width at pectoral fins | BW | |
| Inter-pelvic fin distance | IPvFD | |
| Dorsal-fin base length | DFL | |
| Adipose-fin base length | AdFL | |
| Anal-fin base length | AFL | |
| Caudal-fin base length | CFL | |
| Insertion of dorsal fin to insertion of adipose fin | DF-ADF | |
| Insertion pectoral fin to insertion of pelvic fin | PcF-PvF | |
| Insertion pelvic fin to insertion of anal cavity | PvF-A | |
| Anal cavity to insertion of anal fin | A-AF | |
| Posterior insertion of anal fin to origin of caudal fin | pAF-CF | |
| Posterior insertion of adipose fin and caudal fin | pAdF-CF | |
| Insertion of dorsal fin to operculum end | DF-O | |
| Insertion of dorsal fin to insertion of pectoral fin | DF-PcF | |
| Insertion of dorsal fin to insertion of pelvic fin | DF-PvF | |
| Insertion of adipose fin to insertion of pectoral fin | AdF-PcF | |
| Insertion of adipose fin to insertion of pelvic fin | AdF-PvF | |
| Insertion of adipose fin to insertion of anal fin | AdF-AF | |
| Posterior insertion of adipose fin to posterior insertion of anal fin | pAdF-pAF | |
| Maxillary-barbel length | MB | |
| Outer-mental-barbel length | OMB | |
| Inner-mental-barbel length | IMB | |
| Meristic | Dorsal-fin rays | DFr |
| Pectoral-fin rays | PcFr | |
| Pelvic-fin rays | PvFr | |
| Anal-fin rays | AFr | |
| Caudal-fin upper lobe rays | UCFr | |
| Caudal-fin lower lobe rays | LCFr |
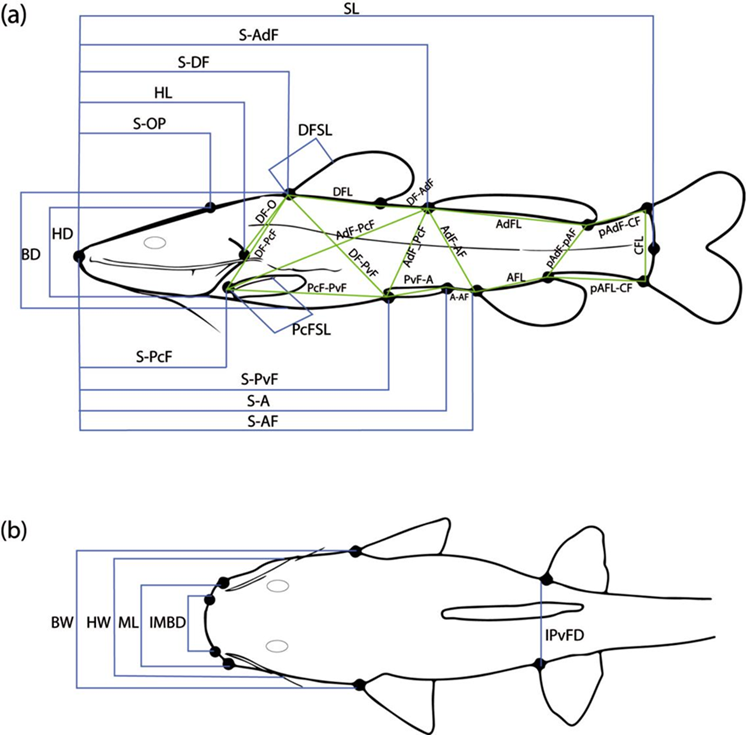
Figure 2 Diagrams displaying linear measurements used to capture overall body shape in lateral (a) and dorsal (b) profiles. Measurement abbreviations are listed in Table 3.
For GM analyses, we employed homologous landmarks of the types 1 and 2, as well as homologous semilandmarks. Type-1 landmarks are those clearly defined by a structure or insertion (e.g., pectoral-fin insertion), whereas those of the type 2 are defined more ambiguously, often as inflection points in curved structures (e.g., posterior margin of the operculum). On the other hand, semilandmarks refer to points along a contour used to represent homologous curves or surfaces (Bookstein, 1997; Gunz et al., 2005). To reduce the criterion of bending energy of the thin-plate spline surface, semilandmarks are allowed to slide along pre-determined vectors (sliders). After sliding, landmarks and semilandmarks can be treated the same way in subsequent statistical analyses (Gunz et al., 2005). The sample for lateral GM (lGM) analysis consisted of 135 specimens: the 129 specimens listed in Table 1 plus the abovementioned 6 from historical collections. Specimens were photographed post-mortem but prior to preservation (except for the 6 ethanol-preserved from historical collections) in a standardized lateral view indicating 15 homologous landmarks (LMs): 14 type 1 (LMs 1-14; mostly located at the insertion of structures) and 1 type 2 (LM 15; posteriormost point of the operculum). Additionally, 7 equidistant semilandmarks (LMs 16-22) were used to capture variation in the dorsal cephalic profile, from the anteriormost tip of the head to the insertion of the dorsal fin (Fig. 3a). For dorsal GM (dGM) analysis, we used a total of 8 landmarks -7 type 1 (LMs 1-7; mostly insertion points) and 1 type 2 (LM 8; posteriormost point of occipital process)- from a smaller subset consisting of 114 specimens but with representation of all sampled localities/populations (Fig. 3b). Just as with meristic and linear trait data, landmark selection was informed by consideration of taxonomically important characters reported in previous studies (Hernández et al., 2015; Silfvergrip, 1996). Homologous landmarks and semilandmarks were digitized with the computer programs tpsDig2 (v.2.31) and tpsUtil (v.1.70), respectively (Rohlf, 2015). The sliders for each semilandmark were defined as tangent vectors to the outline in the position of the point. This tangent was defined as the chord between the previous and next points to the semilandmark along the contour. Shapes were aligned using generalized Procrustes analysis as implemented in the software tpsRelw (v. 1.67) (Rohlf, 2015), removing non-shape variation due to specimen size, location, and orientation. Overall patterns of shape variation were visualized with a PCA paired with thin-plate spline analysis (Bookstein, 1989) to generate warp grids for displaying shape deformations at extremes of principal component (PC) axes relative to mean shape. Relative warps (RWs) analysis was carried out in the software tpsRelw (v. 1.67) (Rohlf, 2015). We tested for measurement error (Bailey & Byrnes, 1990) due to digitizing by means of a one-way Procrustes ANOVA based on 2 alternative digitization schemes for a sample of 30 individuals (1,000 randomized residual permutations) with the function ‘procD.lm’ of the R package geomorph (Adams & Otárola-Castillo, 2013). The resultant estimates of repeatability and measurement error were 98.75% and 1.25%, respectively, and the “individual” main effect was highly significant (p < 0.001). These results imply that the variation among individuals greatly exceeds the measurement error due to digitizing, confirming that variation in residuals was not due to digitizing error but to biological variation. To further investigate group structure and differentiation in multivariate body shape data, a Canonical Variate Analysis (CVA) was performed on both lateral- and dorsal-view Procrustes shape coordinates. By finding shape features that maximize the discrimination among pre-defined groups (relative to the variation within groups), CVA is often used for assessment of group separation and thus to support the identification of distinct species. The statistical significance of pairwise differences in mean shapes was assessed by means of permutation tests based on both Procrustes and Mahalanobis distances and 10,000 permutations per test. CVA and permutation tests were conducted with MorphoJ (v. 1.07) (Klingenberg, 2011).
Results
For all 3 datasets, the best-fit substitution model was the general time-reversible (GTR) with rate heterogeneity among sites modeled by the gamma distribution (Γ) and a proportion of invariant sites (I). The phylogenies resulting from analysis of the COI, CYB, and COI +CYB datasets are presented in Figure 4. In all phylogenies, Mexican species of the “R. laticauda-group” (Weber & Wilkens, 1998; Wilkens, 2001) are resolved as forming a well-supported clade -demarcated by a gray dotted line- that also includes the Costa Rican/Nicaraguan Rhamdia nicaraguensis. Previously undocumented populations from non-type-locality caves in the Sierra de Zongolica, Veracruz (R. cf. zongolicensis), are also resolved as nested within the abovementioned clade, closely related to -but not reciprocally monophyletic with- R. zongolicensis. Regardless of dataset, the phylogenetic placement of troglobitic species (including R. cf. zongolicensis) renders R. laticauda non-monophyletic. The monophyly of R. laticauda is similarly challenged by the placement of the epigean R. nicaraguensis and R. parryi. On the other hand, the monophyly of troglobitic species is not contested by the phylogeny resulting from analysis of the dataset in which multiple individuals of each of them were sampled (i.e., CYB) (Fig. 4b). All phylogenies reveal a modest level of lineage differentiation -as implied by branch lengths- in the troglobitic R. laluchensis and R. macuspanensis. Conversely, the degree of lineage divergence between the troglobitic R. reddelli and R. zongolicensis (including R. cf. zongolicensis) and R. laticauda samples from Mexico (particularly from the Papaloapan basin in Veracruz) appears to be negligible.
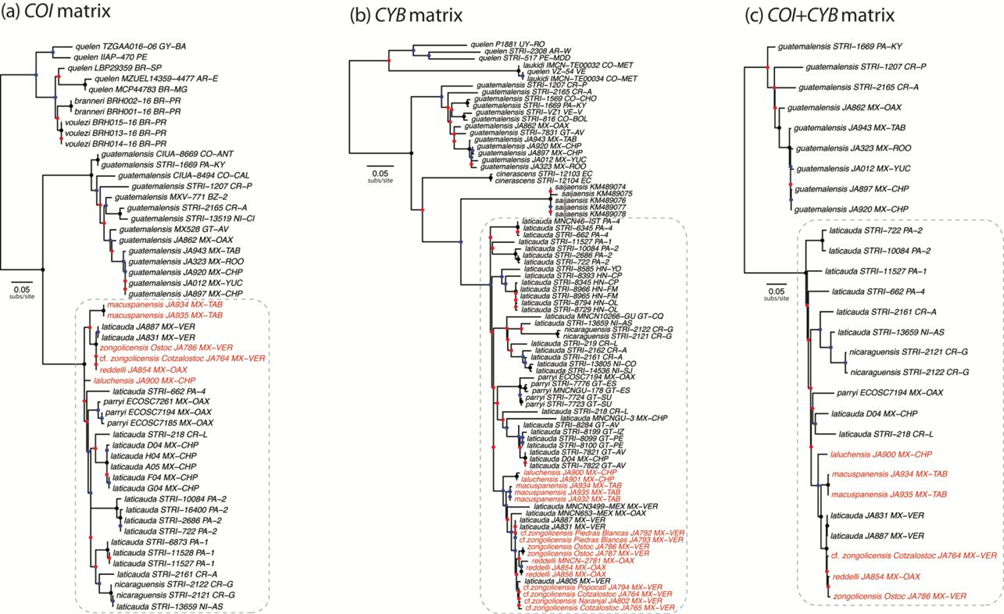
Figure 4 Phylogenetic relationships among sampled Rhamdia species based on (a) the COI matrix, (b) the CYB matrix, and (c) the concatenated COI +CYB matrix. Colored circles on nodes indicate degree of clade support as determined by bootstrap values: black > 0.95, 0.95 ≥ blue ≥ 0.75, red < 0.75. Terminal names as follow: species epithet + voucher + country and province/state (abbreviated using ISO 3166 codes). Terminals corresponding to samples from troglobitic species/populations in red. Outgroup taxon (Pimelodella chagresi) not shown. In each case, the dashed rounded rectangle indicates a well-supported clade formed by members of the “R. laticauda-group” inclusive of R. nicaraguensis.
The resulting COI UPGMA dendrogram displaying clustering of samples according to the (K2P-corrected) genetic distances between them is presented in Figure 5. The greatest divergences are those between R. guatemalensis samples and the remaining sampled Rhamdia species (up to 12.5%). Genetic distances among species of the “R. laticauda-group” do not exceed 4% for any pairwise comparison, with the majority being under 2.5%. The most COI -divergent of the troglobitic species is R. macuspanensis, at ~4% from the remaining troglobitic forms, which do not differ among them in more than ~ 2%. Barcodes of R. reddelli and R. zongolicensis (including R. cf. zongolicensis from the Cotzalostoc cave) are effectively identical, and barely divergent (< 1%) from those of R. laticauda samples from the Papaloapan basin.
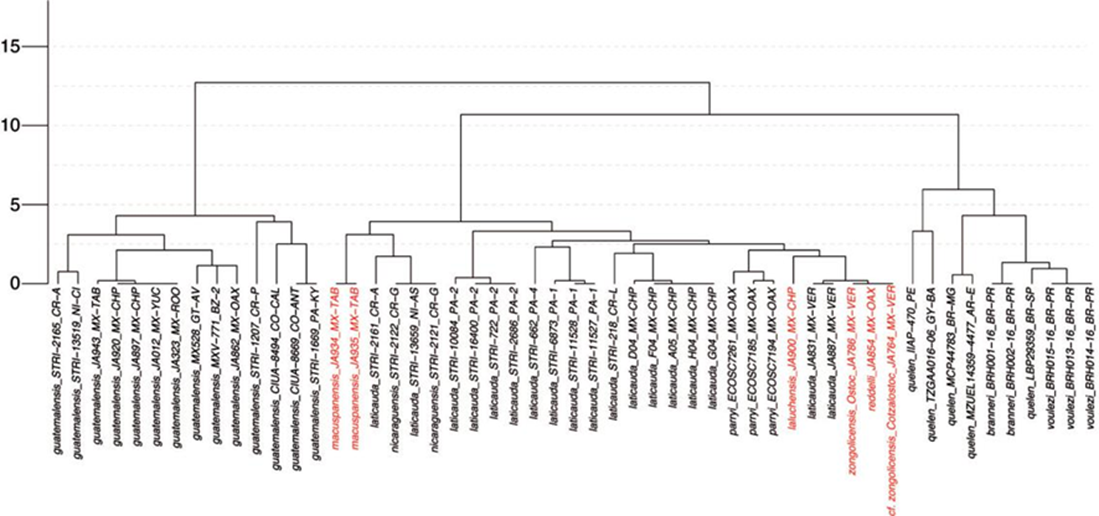
Figure 5 UPGMA dendrogram displaying clustering of samples according to the (K2P-corrected) COI genetic distances. Vertical axis indicates percentage of sequence divergence. Samples from troglobitic species/populations in red.
A broad spectrum of variation in the degree of troglomorphism (i.e., eye reduction and depigmentation) was observed in most troglobitic species, including populations from non-type-locality caves in the Sierra de Zongolica (Fig. 6). In the case of R. zongolicensis, none of the sampled individuals displayed complete eye loss and depigmentation (Fig. 6b). Notably, our sample of R. macuspanensis did not exhibit varying degrees of troglomorphism, but instead only individuals either fully or non-troglomorphic; that is, syntopy of cave and surface forms (Figs. 6c, 7). Taxonomic designation of the non-troglomorphic R. macuspanensis specimen (CNPE-IBUNAM 23814, JA935) was based on multiple sources of evidence such as morphometric and meristic traits, genetics (COI barcodes), and geographic distribution (co-occurrence in the type-locality cave).

Figure 6 Specimens of cave-dwelling Rhamdia in dorsal view showing the spectrum of intraspecific variation in troglobitic characters (eye reduction and depigmentation) in A) R. reddelli, B) R. zongolicensis, C) R. macuspanensis, D) R. laluchensis, E) R. cf. zongolicensis (Sumidero de Cotzalostoc), and F) R. cf. zongolicensis (Sótano de Popocatl).
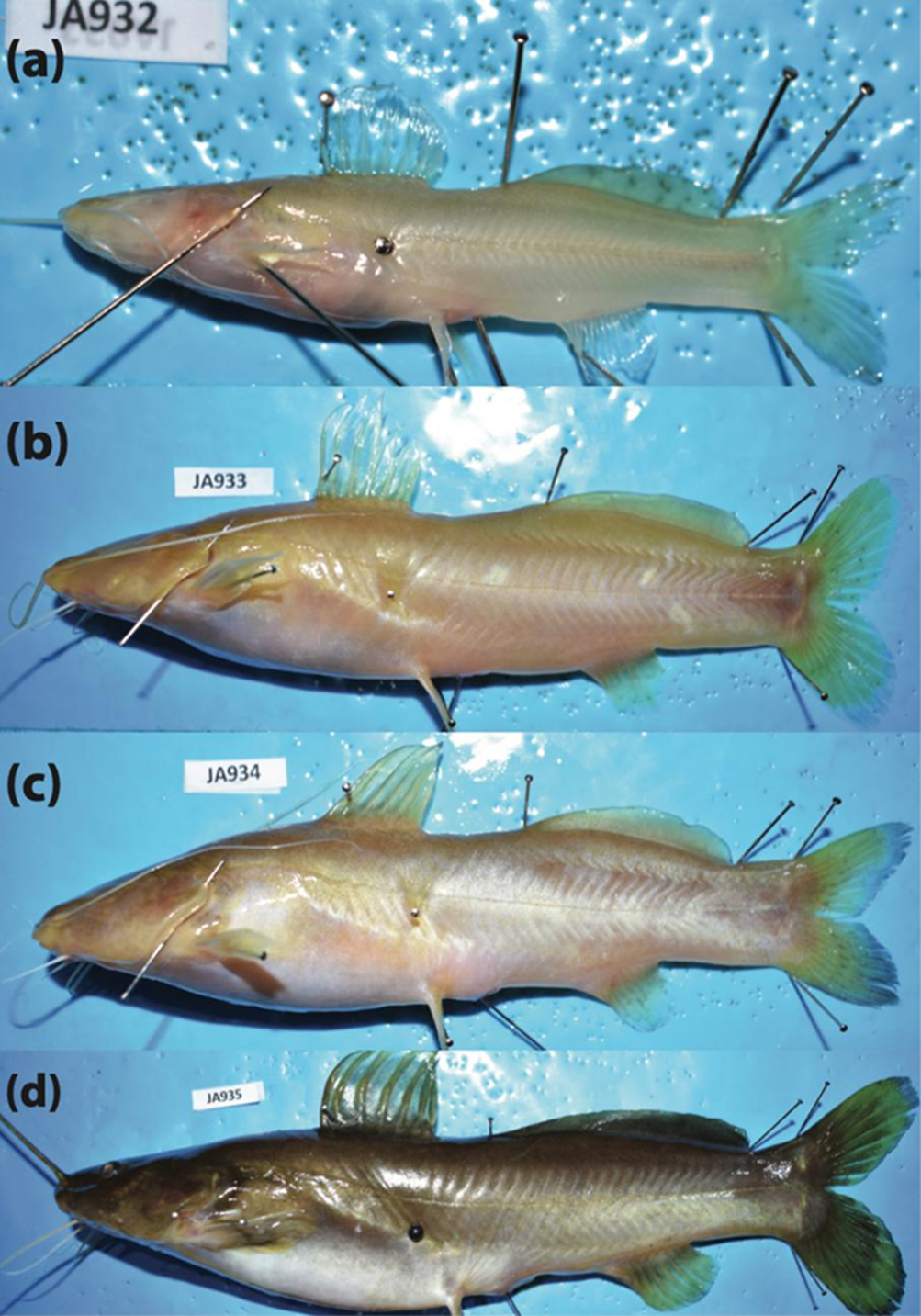
Figure 7 Specimens of R. macuspanensis in lateral view showing syntopy of troglobitic and epigean forms at its type-locality cave, Grutas de Agua Blanca (Tabasco).
The distribution of meristic counts in troglobitic forms (all 4 described species plus 2 undescribed populations/ species) and the epigean species R. laticauda and R. guatemalensis is presented in Table 4. The PCA on linear data resulted in 37 PCs that explain 100% of the variation, with only the first 3 individually accounting for more than 10%, and together explaining 52.6% of the overall variance. A graphical representation of these results is presented in Figure 8. The first PC is primarily associated with variation in barbel length, being the maxillary barbels those exhibiting the greatest variation. Despite the considerable overlap among species/populations along this PC, it can be observed that samples from the epigean R. guatemalensis and the hypogean R. macuspanensis cluster at the upper half (implying longer barbels), whereas those from the epigean R. laticauda and the hypogean R. cf. zongolicensis are mostly confined to the lower half (implying shorter barbels). Samples from the remaining species (R. reddelli, R. zongolicensis, and R. laluchensis) are highly overlapping and cluster at intermediate values along this gradient of variation. The second PC is primarily associated with variation in body depth. With the exception of R. macuspanensis, which displays a relatively robust body more akin to that of epigean species, samples from troglobitic species/populations cluster primarily towards the negative side of this axis, meaning that they have considerably slenderer bodies. Lastly, the third PC is mostly associated with variation in the length of the interdorsal space. The observed pattern implies that R. guatemalensis is the most divergent in this respect, having the shortest interdorsal space with respect to R. laticauda and the troglobitic forms, which together display almost complete overlap and are therefore indistinguishable as far as this character is concerned. Despite the apparent lack of clear differentiation among species/populations implied by the general patterns of morphometric variation uncovered via PCA (except for R. guatemalensis and R. macuspanensis) (Fig. 8), MANOVA results indicate that in fact there are statistically significant differences among groups, a finding that holds across most pairwise comparisons (Tables 5, 6). Notably, R. laticauda was found to be significantly different in overall body shape from all troglobitic species/populations. In contrast, no significant differences were found between R. reddelli and R. zongolicensis.
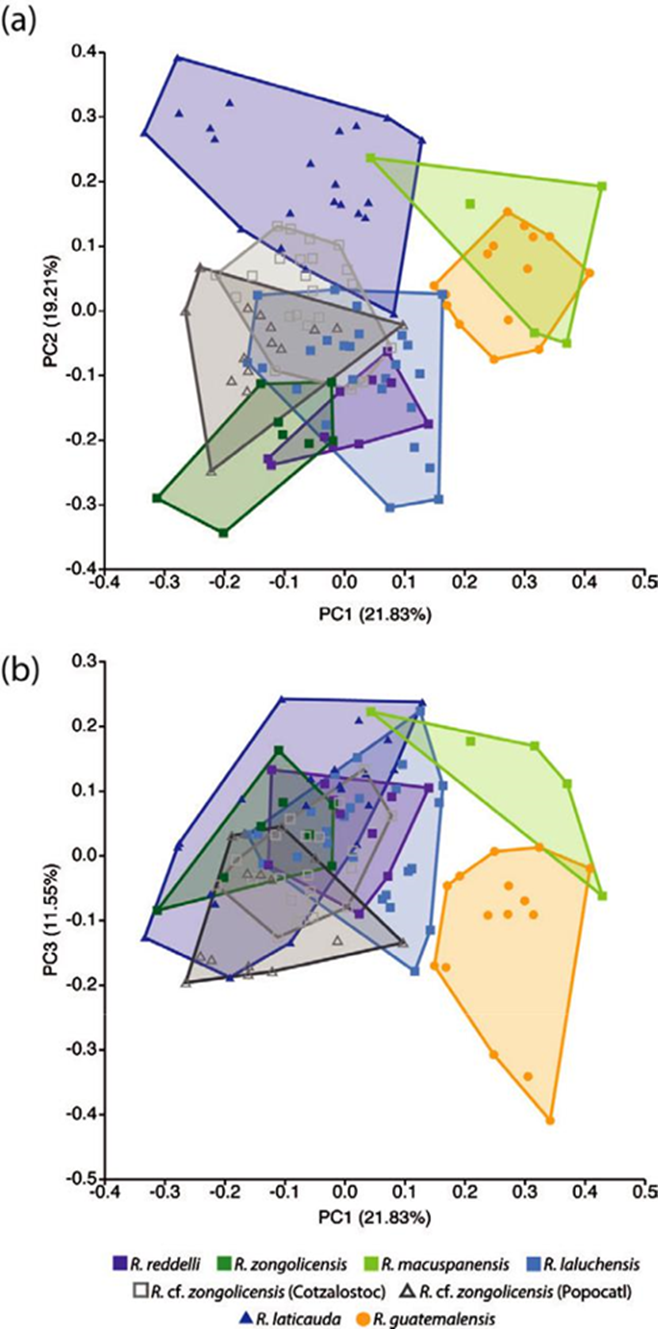
Figure 8 Results of Principal Component Analysis (PCA) of overall body shape variation based on linear data.
Table 4 Summary of the variation in meristic trait data registered for all 4 troglobitic species, 2 undescribed cave-dwelling populations (R. cf. zongolicensis 1 = Sumidero de Cotzalostoc; R. cf. zongolicensis 2 = Sótano de Popocatl), and the epigean R. laticauda and R. guatemalensis. Total number of individuals in parenthesis. Meristic formula: spine counts in Roman numerals; (soft) ray counts in Arabic. Abbreviations: DFr = dorsal-fin rays; PvFr = pelvic-fin rays; PcFr = pectoral-fin rays; AFr = anal-fin rays; UCFr = upper caudal-fin rays; LCFr = lower caudal-fin rays.
| Species | DFr | PvFr | PcFr | AFr | UCFr | LCFr |
| R. reddelli (10) | I 6 (10) | 6 (10) | I 8 (2), I 9 (6), I 10 (2) | 11 (2), 12 (1), 13 (3), 14 (2), 15 (1), 16 (1) | 8 (10) | 9 (2), 10 (8) |
| R. zongolicensis (8) | I 5 (1), I 6 (7) | 6 (8) | I 8 (1), I 9 (7) | 11 (2), 12 (2), 13 (3), 14 (1) | 7 (1), 8 (7) | 10 (7), 11(1) |
| R. macuspanensis (5) | I 6 (5) | 6 (5) | I 8 (3), I 10 (2) | 10 (1), 11 (2), 13 (1), 14 (1) | 8 (5) | 9 (1), 10 (4) |
| R. laluchensis (25) | I 6 (25) | 6 (25) | I 7 (1), I 8 (1), I 9 (10), I 10 (11), I 11 (2) | 11 (6), 12(13), 13(4), 14(2) | 7 (2), 8(22), 9(1) | 9 (6), 10 (18), 11 (1) |
| R. cf. zongolicensis1 (22) | I 6 (22) | 4 (1), 6 (21) | I 8 (10), I 9 (11), I 10 (1) | 11 (8), 12 (11), 13 (2), 14 (1) | 8 (21), 9 (1) | 9 (3), 10 (18), 11 (1) |
| R. cf. zongolicensis2 (15) | I 6 (15) | 6 (15) | I 8 (4), I 9 (9), I 10 (2) | 12 (2), 13 (4), 14 (5), 15 (3), 16 (1) | 7 (3), 8 (12) | 8 (2), 9 (7), 10 (6) |
| R. laticauda (22) | I 6 (20), I 7 (2) | 6 (22) | I 7 (1), I 8 (9), I 9(12) | 9 (1), 10 (3), 11 (10), 12 (6), 13 (2) | 6 (1), 7 (4), 8 (16) | 7 (2), 10 (20) |
| R. guatemalensis (15) | I 6 (15) | 6 (15) | I 7 (1), I 8 (12), I 9 (2) | 10 (1), 11 (5), 12 (8), 13 (1) | 8 (14), 9 (1) | 10 (14), 11(1) |
Table 5 Results of MANOVA on linear trait data.
| Test statistic | df1 | df2 | F | p-value | |
| Wilks’ lambda | 0.06074 | 21, | 322.2 | 25.4 | < 0.0001 |
| Pillai’s trace | 1.707 | 21, | 342 | 21.5 | < 0.0001 |
Table 6 P-values for all possible pairwise morphometric comparisons between the Rhamdia species/populations investigated in this study (Hotelling’s T 2 test; α = 0.05). Values with asterisk indicate no significant differences (i.e., p-value > α). R. cf. zongolicensis 1 = Sumidero de Cotzalostoc; R. cf. zongolicensis 2 = Sótano de Popocatl.
| R. | R. | R. | R. | R. | R. | R. cf. | |
| guatemalensis | laticauda | laluchensis | macuspanensis | reddelli | zongolicensis | zongolicensis 1 | |
| R. guatemalensis | |||||||
| R. laticauda | < 0.0001 | ||||||
| R. laluchensis | < 0.0001 | < 0.0000 | |||||
| R. macuspanensis | 0.0421 | 0.0002 | < 0.0000 | ||||
| R. reddelli | < 0.0001 | < 0.0001 | 4.1082* | 0.0023 | |||
| R. zongolicensis | < 0.0001 | < 0.0001 | 0.0057 | 0.0014 | 2.2947* | ||
| R. cf. zongolicensis 1 | < 0.0001 | < 0.0001 | 0.0009 | 0.0001 | 0.0009 | 0.0001 | |
| R. cf. zongolicensis 2 | < 0.0001 | < 0.0001 | 0.0011 | < 0.0001 | 0.029 | 0.0638* | 0.0668* |
The PCA of lateral-view body shape in the sampled specimens indicates that 40 principal components/RWs explain 100% of the variation, with the first 2 explaining 41.92% of the overall shape variance. The largest component of shape variation (RW1), accounting for 25.76% of the total variance, is associated primarily with changes in body depth (at the level of pelvic fin) and, to a lesser extent, with head length (Fig. 9a). Except for R. macuspanensis, samples from troglobitic species/populations cluster towards the positive side of RW1 (i.e., slenderer, longer-headed bodies), whereas samples from the epigean species R. guatemalensis and R. laticauda scatter on the negative side (i.e., deeper, shorter-headed bodies). The second-largest component of shape variation (RW2) explains 16.66% of total variance and is associated mainly with changes in interdorsal space and, to a smaller degree, with head depth (Fig. 9a). Therefore, individuals located on the positive side of RW2 display a longer interdorsal space coupled with a shorter adipose fin, as well as a slenderer head compared to those on the negative side of this RW. Troglobitic species display considerable overlap on this axis, with most of the clustering at intermediate values (near zero), and with R. laluchensis displaying the widest range of variation (possibly due to having the largest sample size). Although R. laticauda displays the largest values along this axis (i.e., largest interdorsal space and slenderest head), these partially overlap with those of troglobitic forms, particularly at the lower half of the range. On the other hand, the PCA of dorsal-view body shape resulted in 12 RWs that explain 100% of the variation, with the first 2 explaining 66.07% of the overall variance. In this case, the largest component of variation (RW1) explains most of the total variance (52.34%) and is associated with variation in head size and shape and in the relative position of the pectoral and pelvic fins (Fig. 9b). Specifically, extreme values along this axis describe either individuals with longer and sharper heads and paired fins closer together (on the negative side) or with shorter and blunter heads and paired fins more apart (on the positive side). With the exception of R. macuspanensis, which displays a broad range of values that extend on both negative and positive sides of this axis, samples from described troglobitic species cluster on the negative side and therefore have comparatively larger heads and paired fins closer together. On the other hand, undescribed troglobitic populations from the Sierra de Zongolica (Popocatl and Cotzalostoc) display intermediate values along this axis, while simples from R. laticauda occur mostly on the positive side. Variation in the second-largest component of dorsal-view shape variance (RW2; 13.71%) is mainly explained by the shape of the trunk, particularly as it relates to the position of the pelvic fins, with fishes on the positive side of the axis displaying posteriorly narrower bodies (Fig. 9b). Except for R. guatemalensis, whose individuals cluster narrowly towards the central region, sampled populations display a broad and highly overlapping range of variation along this axis, implying that this shape component is not particularly useful to morphologically differentiate species. The results from CVA on both lateral and dorsal profiles are basically equivalent to the abovementioned general patterns of morphological variation implied by PCA (Fig. 10). Lateral-shape CVA indicates that 7 canonical variates (CV) explain 100% of the variation, the first 2 of which account for 63.15% of the overall shape variance (Fig. 10a). The largest component of lateral shape variation (CV1; 35.51% of total variance) is primarily associated with changes in body depth and head length. Lower values on this axis indicate deeper bodies and shorter heads (such as in R. guatemalensis), while higher values specify slenderer bodies with larger heads (such as in most troglobitic species) (Fig. 10a). The second-largest component of lateral shape variation (CV2; 27.64% of total variance) is associated mainly with changes in interdorsal space. For the most part, troglobitic species occur at intermediate values along this axis, between R. guatemalensis (at the positive extreme) and R. laticauda (at the negative extreme) (Fig. 10a). On the other hand, CVA of dorsal shape data resulted in 7 CVs that explain 100% of the variation, of which the first 2 account for 80.59% of the overall shape variance (Fig. 10b). The largest component of dorsal shape variation (CV1; 67.59% of total variance) is mainly related to head size and body width at pelvic-fin origin. Most troglobitic species and populations fall on the positive side of this axis (i.e., have longer heads and narrower bodies) with considerable overlap, while the epigean species and the troglobitic R. macuspanensis are mostly restricted to the negative side (i.e., have smaller heads and thicker bodies) (Fig. 10b). The second-largest component of dorsal shape variation (CV2; 13% of total variance) is related to changes in head and trunk width. The substantial overlap among species along this axis, however, entails no discernible pattern of differentiation based on this shape attribute (Fig. 10b). Overall, with few exceptions, all pairwise comparisons of lateral shape resulted in statistically significant differences (Table 7).
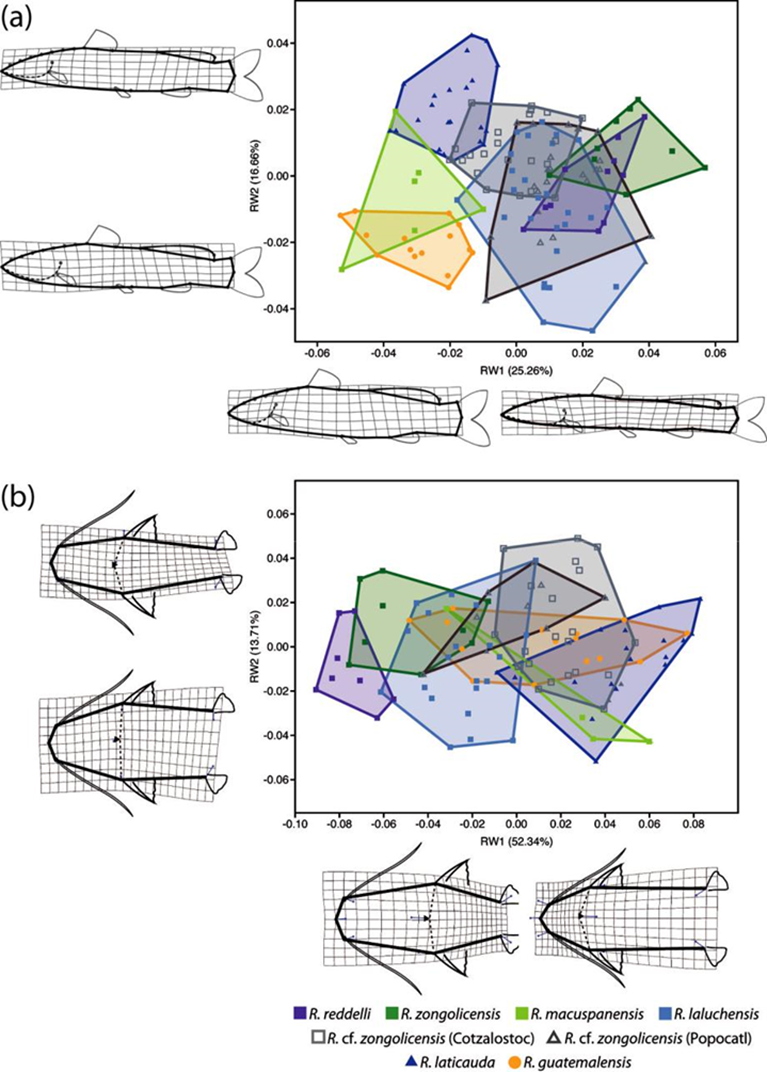
Figure 9 Results from Relative Warps (RWs) analysis of overall body shape variation based on landmark data. Deformation grids display the nature of shape change across major RWs in lateral (a) and dorsal (b) profiles.
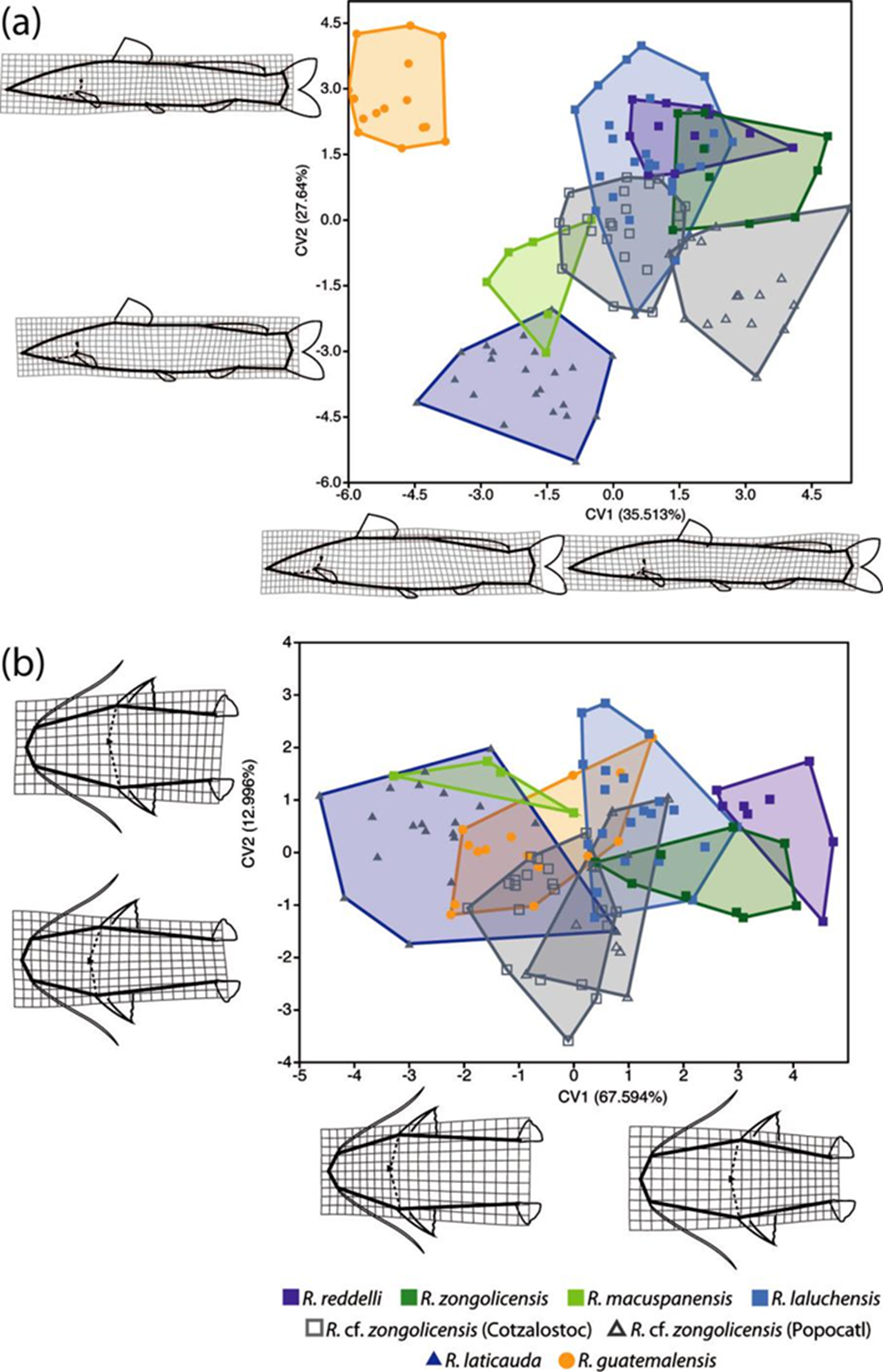
Figure 10 Results from Canonical Variate Analysis (CVA) of overall body shape variation based on landmark data in lateral (a) and dorsal (b) profiles.
Table 7 P-values of permutation tests (α = 0.0001) of pairwise differences in mean body shape (lateral profile) based on Procrustes (standard typeface) and Mahalanobis (bold) distances. Values with asterisk indicate no significant differences (i.e., p-value > α). R. cf. zongolicensis 1 = Sumidero de Cotzalostoc; R. cf. zongolicensis 2 = Sótano de Popocatl.
| R. guatemalensis | R. laticauda | R. laluchensis | R. macuspanensis | R. reddelli | R. zongolicensis | R. cf. zongolicensis1 | R. cf. zongolicensis2 | |
| R. guatemalensis | - | < 0.0001 | < 0.0001 | < 0.0001 | < 0.0001 | < 0.0001 | < 0.0001 | < 0.0001 |
| R. laticauda | < 0.0001 | - | < 0.0001 | < 0.0001 | < 0.0001 | < 0.0001 | < 0.0001 | < 0.0001 |
| R. laluchensis | < 0.0001 | < 0.0001 | - | < 0.0001 | 0.0001 | < 0.0001 | < 0.0001 | 0.0011* |
| R. macuspanensis | < 0.0001 | < 0.0001 | < 0.0001 | - | 0.0002* | < 0.0001 | < 0.0001 | |
| R. reddelli | < 0.0001 | < 0.0001 | < 0.0001 | < 0.0001 | - | 0.036* | < 0.0001 | < 0.0001 |
| R. zongolicensis | < 0.0001 | < 0.0001 | < 0.0001 | 0.0002* | 0.0049* | - | < 0.0001 | < 0.0001 |
| R. cf. zongolicensis1 | < 0.0001 | < 0.0001 | < 0.0001 | < 0.0001 | < 0.0001 | < 0.0001 | - | 0.0002* |
| R. cf. zongolicensis2 | < 0.0001 | < 0.0001 | < 0.0001 | < 0.0001 | < 0.0001 | < 0.0001 | < 0.0001 | - |
In contrast, pairwise comparisons of dorsal shape resulted in a smaller number of significantly different species pairs (Table 8). Notably, these results imply that R. zongolicensis does not differ significantly in lateral profile from either R. reddelli or R. macuspanensis, and that R. macuspanensis does not differ significantly in dorsal profile from any of the remaining species/populations investigated.
Table 8 P-values of permutation tests (α = 0.0001) of pairwise differences in mean body shape (dorsal profile) based on Procrustes (standard typeface) and Mahalanobis (bold) distances. Values with asterisk indicate no significant differences (i.e., p-value > α). R. cf. zongolicensis 1 = Sumidero de Cotzalostoc; R. cf. zongolicensis 2 = Sótano de Popocatl.
| R. guatemalensis | R. laticauda | R. laluchensis | R. macuspanensis | R. reddelli | R. zongolicensis | R. cf. | R. cf. | |
| zongolicensis1 | zongolicensis2 | |||||||
| R. guatemalensis | - | 0.001* | < 0.0001 | 0.1707* | < 0.0001 | 0.0002* | 0.0501* | 0.1051* |
| R. laticauda | 0.0001 | - | < 0.0001 | 0.1155* | < 0.0001 | < 0.0001 | < 0.0001 | < 0.0001 |
| R. laluchensis | < 0.0001 | < 0.0001 | - | 0.0031* | < 0.0001 | 0.0002* | < 0.0001 | 0.0018* |
| R. macuspanensis | 0.0879* | 0.206* | 0.0004* | - | 0.0013* | 0.0054* | 0.0658* | 0.0279* |
| R. reddelli | < 0.0001 | < 0.0001 | 0.0001 | 0.0011* | - | 0.0011* | < 0.0001 | < 0.0001 |
| R. zongolicensis | < 0.0001 | < 0.0001 | 0.0001 | 0.0007* | 0.0001 | - | < 0.0001 | 0.0022* |
| R. cf. zongolicensis1 | < 0.0001 | < 0.0001 | < 0.0001 | 0.006* | < 0.0001 | < 0.0001 | - | 0.0465* |
| R. cf. zongolicensis2 | 0.0001 | < 0.0001 | 0.0031* | 0.0025* | < 0.0001 | 0.0083* | 0.0144* | - |
Discussion
Although our focus was the pattern of relationships of Mexican troglobitic Rhamdia in the context of the Middle American radiation of the genus, our results broadly conform with previous molecule-based hypotheses of relationships inferred for non-troglobitic Rhamdia across the entire geographic range of the genus (Hernández et al., 2015; Perdices et al., 2002) (Fig. 4). The phylogenetic evidence we present here implies that Mexican troglobitic species are part of a clade that also includes the epigean species R. laticauda, R. parryi, and R. nicaraguensis. The inferred phylogenetic placement of troglobitic forms, as well as that of R. parryi and R. nicaraguensis, however, renders R. laticauda irreconcilably non-monophyletic. Under this phylogenetic framework, continued recognition of R. laticauda as currently delimited is problematic and undesirable. Notwithstanding, our phylogenetic results (CYB phylogeny) also fail to refute the monophyly of all recognized troglobitic species (with respect to R. laticauda), while revealing a modest level of lineage differentiation in R. laluchensis and R. macuspanensis. Granted, such a degree of lineage distinction does not necessarily entail interspecific differentiation, for it is also consistent with intraspecific geographic genetic structuring under the hypothesis that troglobitic species are in fact cave-adapted populations of R. laticauda. Comparatively larger mtDNA divergences between epigean and hypogean populations, have been reported for the Mexican tetra A. mexicanus (Ornelas-García et al., 2008), and yet both cave and surface forms are presently regarded as a single species by many authors (Gross, 2012; Hausdorf et al., 2011; Miller, 2005; Strecker et al., 2012). Thus, our phylogenetic findings offer support to the hypothesis that Mexican troglobitic species of Rhamdia, as well as the epigean R. parryi and R. nicaraguensis, effectively represent lineages of the more widespread species R. laticauda.
Whether these findings warrant a taxonomic overhaul, however, deserves further scrutiny. Reciprocal monophyly is a property that lineages acquire as they separate and diverge from one another, and therefore it is widely considered as evidence in support for the process of speciation (de Queiroz, 2007). Although a number of authors have argued for the existence and recognition of paraphyletic species (e.g., Crisp & Chandler, 1996; Kizirian & Donnelly, 2004), paraphyly of “ancestral” species, while certainly a possibility (particularly in speciation by peripheral isolation), would only be transient under the assumption that species ultimately evolve into monophyletic lineages (de Queiroz, 2007). Accordingly, our phylogenetic results imply that Mexican troglobitic forms of Rhamdia represent either highly localized, “exclusive” cave-dwelling populations of the widespread and primarily “non-exclusive” epigean species R. laticauda (“exclusiveness” sensu Wiens & Penkrot, 2002) or recently diverged species at very early stages of their speciation from R. laticauda. We recognize that our genetic data might be insufficient to unambiguously distinguish between these 2 contrasting hypotheses. Therefore, until these alternative scenarios can be properly tested, caution should be exercised when drawing taxonomic conclusions based solely on the phylogenetic evidence presented herein. Our intention is not to prioritize taxonomic concerns -such as the monophyly of binominals- over the recognition of diversity, but to shed light on the evolution and systematics of the genus Rhamdia by testing the species status ascribed to long-recognized Mexican troglobitic populations using multiple lines of evidence, including phylogenetic.
Notably, this is not the first time that taxonomic/ nomenclatural issues involving R. laticauda and closely related species have been raised. In his revision of the genus, Silfvergrip (1996) concluded that the troglobitic species described at the time -R. zongolicensis and R. reddelli- as well as the epigean R. parryi, were junior synonyms of R. laticauda. Although he considered R. nicaraguensis valid and distinct from R. laticauda, the main character he proposed to distinguish them (i.e., shorter adipose fin) had been previously shown to display considerable overlap between the 2 species (Bussing, 1987; Silfvergrip, 1996, p. 92). Similarly, in the only previous study that sampled a Mexican troglobitic species of Rhamdia for phylogenetic analysis, the authors concluded that “preserving the species status of R. reddelli would suggest that more than 20 additional species should be recognized in the R. laticauda clade alone if a taxonomic goal is to fairly represent evolutionary history” (Perdices et al., 2002, p. 182). It should be noted that, based on their resultant phylogeny, the same conclusion would apply to R. parryi and R. nicaraguensis as it applies to R. reddelli (Perdices et al., 2002, p. 177, Fig. 2).
According to our results, among the troglobitic species, R. reddelli and R. zongolicensis are by far the most closely related phylogenetically. This conforms with the taxonomic assessment of Miller (2005) -who regarded them as synonyms- and with the expectations from geography. The type-locality caves of these 2 species are in relative proximity to one another (~ 30 km) and are part of the same hydrological basin (Papaloapan). Along this line of reasoning, our finding that R. laticauda samples from the Papaloapan are the most closely related to R. reddelli and R. zongolicensis (including R. cf. zongolicensis) is not only unsurprising but also supportive of the hypothesis that ancestral populations of R. laticauda from the region colonized hypogean habitats resulting in the evolution of the cave-adapted troglobitic forms present in the Sierra de Zongolica (Veracruz) and neighboring karstic systems (Cueva del Nacimiento del Río San Antonio, Oaxaca). The timing and other specifics about this colonization, however, have yet to be determined. Our findings also imply that R. laluchensis appears to be the earliest divergent of the Mexican troglobitic species, but details on the history of colonization of the Sótano de La Lucha (Chiapas) - as well as of the Grutas de Agua Blanca (Tabasco) in the case of R. macuspanensis- are presently unknown. Notwithstanding, our phylogenetic results suggest that the known diversity of troglomorphic Rhamdia in southern Mexico can be explained by at least 3 cave colonization events by the epigean species R. laticauda leading to the establishing of troglomorphic populations in caves of Chiapas (R. laluchensis), Tabasco (R. macuspanensis), and Veracruz/Oaxaca (R. zongolicensis and R. reddelli), respectively. A larger geographic sampling of epigean populations will be required to shed further light on the apparently complex history of cave colonization by Rhamdia in southern Mexico.
Traditional DNA barcoding species identification relies on the use of thresholds set to differentiate between intraspecific variation and interspecific divergence (Hebert et al., 2003, 2004). In fishes, the proposed ~ 3% COI sequence divergence heuristic threshold for conspecifics (Ward, 2009) appears to hold across a broad range of teleost lineages (Arroyave et al., 2019; Decru et al., 2016; Lara et al., 2010; Lowenstein et al., 2011; Pereira et al., 2011, 2013; Valdez-Moreno et al., 2009). Our phenetic analysis of COI sequence variation in Rhamdia uncovered unexpectedly low divergences (mostly < 3%) among species of the “R. laticauda-group” inclusive of R. nicaraguensis, effectively impeding unambiguous identification and distinction of most troglobitic species via DNA barcoding. Even the largest divergences among samples from this multispecies group (~ 4%) pale in comparison with the divergences between R. laticauda and demonstrably different species such as R. guatemalensis (~ 12%) or the cis-Andean R. quelen (~ 10%). The uncovered patterns of DNA barcode variation in Rhamdia are therefore consistent with the hypothesis that currently recognized troglobitic species effectively correspond to cave-adapted lineages/populations of the more widespread, epigean R. laticauda. Because of the limitations of the method, however, in and of themselves these results should not be interpreted as sufficient evidence of conspecificity among troglobitic forms.
In support for the recognition of R. reddelli and R. zongolicensis, Wilkens argued that “considerable genetic divergences exist between them” and therefore “morphological similarity is the result of convergent evolution and the cave catfishes should be treated as separate species” (Wilkens, 2001, p. 260). Our results from both phylogenetic and phenetic analyses of comparative mtDNA data, however, imply that the degree and pattern of genetic divergences among Mexican troglobitic populations is considerably lower than the expected for samples belonging to different species (“contra” Wilkens, 2001). Remarkably, some of the evidence brought up by Wilkens to support this claim consisted of unpublished data: “the evolution of separate gene pools is proven by mtDNA-analysis (unpublished data)” (Wilkens, 2001, p. 260). Our own analyses of mtDNA data strongly dispute this statement. The remaining genetic evidence cited by Wilkens to endorse the species status of R. reddelli and R. zongolicensis came from previous work in which laboratory crosses between these species and the epigean R. laticauda resulted in F1s consisting exclusively of individuals from the heterogametic sex (in this case females), conforming to Haldane’s Rule (Wilkens, 1993). Hybrid dysfunction under Haldane’s Rule, however, applies to early stages of speciation (Coyne & Orr, 1989; Haldane, 1922). Furthermore, most research on Haldane’s rule has been conducted on Drosophila, thus making generalizations and extensions to other taxa, especially poorly studied ones and with heterogametic females, suspect at best (Johnson, 2008). While we acknowledge that our results are also consistent with patterns expected at early stages of the speciation process, we believe that current available evidence appears insufficient to assert absolute divergence, reproductive isolation, and lineage differentiation worthy of species status in any form of troglobitic Rhamdia from southern Mexico.
Despite previous reports about the coexistence of individuals with varying degrees of troglomorphism in populations of Mexican troglobitic Rhamdia, the implications and generality of this pattern had not been critically investigated (Mosier, 1984; Sbordoni et al., 1986; Wilkens, 2001). Our qualitative assessment of intraspecific variation in ocular development and degree of pigmentation not only corroborates the aforementioned reports, but also reveals that this phenomenon is more widespread than previously documented, effectively occurring in all cave populations examined (Fig. 6), and even manifested through previously unreported syntopy of epigean and troglobitic forms in R. macuspanensis (Fig. 7). Regarding the abovementioned syntopy of forms, we tentatively attribute the absence of intermediate phenotypes in R. macuspanensis to sampling error (effectively the smallest sample size at n = 5).
Intermediate troglobitic phenotypes have been documented both in the wild and in F1 laboratory crosses between surface and cave forms of the Mexican tetra A. mexicanus, and their existence attributed to either recent divergence from surface populations (i.e., not enough time since hypogean colonization for fixation of troglomorphic traits) or present-day hybridization between cave and surface fish (Wilkens, 2016). All populations of troglobitic Rhamdia investigated in this study live in pools inside caves that are supplied, either perennially or intermittently, by surface water (e.g., streams, surface runoff) and/ or groundwater from karstic springs. Therefore, it is reasonable to assume that individuals from the epigean R. laticauda have the potential to “hybridize” with cave-dwelling fish. Remarkably, complete replacement of a cave-dwelling troglomorphic population by its epigean ancestor has been previously documented in Rhamdia, namely R. quelen from the Cumaca cave in Trinidad, West Indies (Romero et al., 2002). Originally described as Caecorhamdia urichi Norman 1926, this troglomorphic population was subsequently synonymized with R. quelen precisely because of its lack of diagnostic features other than the absence of eyes and depigmentation (Mees, 1974; Silfvergrip, 1996). Varying degree of troglomorphism had been reported for this population since the 1950s and, by the early 2000s, the troglobitic phenotype had been completely substituted with the surface form, possibly prompted by changes in precipitation regimes and the ability of epigean individuals to outcompete troglobitic ones (Romero et al., 2002). Without entirely discounting the possibility of intraspecific phenotypic plasticity (Bilandžija et al., 2020), we hypothesize that the observed clinal variation in troglomorphism in Mexican cave-dwelling species/populations of Rhamdia (Figs. 6, 7) is the result of crossings between epigean and hypogean forms rather than recent divergence followed by only partial fixation of troglomorphism. Further research into this subject (e.g., QTL analysis of genome-wide data), however, is required to properly test this hypothesis and better understand the genetic basis of regressive phenotypic loss in Mexican cave-dwelling Rhamdia.
Of the 6 meristic variables investigated, dorsal-fin and pelvic-fin ray counts were the ones that exhibited the least amount of variation across all species/populations evaluated (6 rays in almost all samples). In contrast, anal-fin ray counts displayed the widest range of variation (9-16), although with most values concentrated at 11-13 rays, and a slight tendency of troglobitic forms to display a comparatively higher ray count. The remaining variables, while displaying more intermediate ranges, still exhibited a tendency to concentrate most records on a single value (i.e., a clearly defined mode). Overall, none of these meristic traits display a non-overlapping distribution with potential to diagnose or distinguish among troglobitic species, nor between these and the epigean R. laticauda. These findings are consistent with a recurrent issue in catfish systematics, that is, the fact that meristic features, while not necessarily uninformative, are plagued by considerable overlap when a significant number of taxa is compared (Silfvergrip, 1996; p. 120).
Although exploratory analysis of body shape variation as implied by linear data (Fig. 8) did not seem to unambiguously differentiate among troglobitic species, as well as between these and the epigean R. laticauda, MANOVA results did support the existence of significant differences among most of the species/populations investigated (Tables 5, 6). Similarly, GM analyses initially suggested a lack of absolute and clear-cut differentiation in overall body shape among troglobitic forms and between them and R. laticauda (Figs. 9, 10), but subsequently demonstrated the existence of statistically significant differences, at least in the lateral-view component of body shape (Table 7). Whether such differences constitute proof of species-level differentiation (vs. intraspecific variation), however, requires further scrutiny. On one hand, statistically significant differences in body shape between troglobitic species/populations and R. laticauda are ultimately phenetic and therefore cannot be unequivocally attributed to historical processes such as speciation. On the other hand, such differentiation appears to be mainly related with phenotypically plastic features associated with life in hypogean habitats (e.g., body depth, head length).
Despite being a character system explaining a considerable proportion of the observed variation in linear trait data (PC1 in Figure 8), barbels are not consistently longer in troglobitic forms of Mexican Rhamdia with respect to epigean species. This finding is certainly contrary to our expectations, since barbel elongation has long being considered a typical constructive adaptation in troglobitic fishes (Romero & Green, 2005; Soares & Niemiller, 2020). Without discounting other possible explanations, this counterintuitive finding could be attributed to the relative recency of the divergence between R. laticauda and hypogean species/populations (none of the cave-dwelling forms has on average shorter barbels than R. laticauda). Furthermore, the presence of comparatively longer barbels in the epigean R. guatemalensis is unsurprising when considering its strong nyctophilia (Arroyave et al., 2020; Wilkens, 2001).
Among the shape attributes that appear to offer some support for the morphological distinction between R. laticauda and currently recognized troglobitic species are body depth (PC2, RW1, and CV1 in Figures 8a, 9a, 10a, respectively) and head size and shape coupled with the relative position of the pectoral and pelvic fins (RW1 and CV1 in Figures 9b, 10b, respectively). While variation in body depth seems potentially useful to discriminate troglobitic species (except R. macuspanensis) from the epigean R. laticauda, this shape attribute is potentially problematic because cave fishes tend to have slenderer bodies than their epigean counterparts due to the fact that caves are often nutrient limited (Langecker, 2000; Venarsky et al., 2014). Notably, our findings that troglobitic species tend to have longer and sharper heads relative to R. laticauda are in agreement with each and every one of the original diagnoses, which also list the presence of a weak and short occipital process as diagnostic (Miller, 1984; Wilkens, 1993; Weber & Wilkens, 1998; Weber et al., 2003). The fact that troglobitic forms have longer heads begs the question of whether head enlargement in troglobitic Rhamdia, just like barbel elongation, constitutes an instance of constructive troglomorphism and convergent adaptation to hypogean habitats, and therefore of questionable taxonomic utility. While typical constructive troglomorphic traits involve improved chemosensory capacities through elongated sensory structures (Jones & Culver, 1989), head enlargement might be driven solely by functional necessity, to accommodate hypertrophied sensory receptors (olfactory, tactile, lateral-line, etc.) and brain centers (Poulson, 1963). Thus, even if only developmentally and mechanically correlated with traits under positive selection in hypogean habitats (enhanced non-visual sensory systems), head enlargement would be an indirect byproduct of selection, and so, effectively a convergent adaptation.
This study shows that the validity of currently recognized troglobitic species of Mexican Rhamdia is unequivocally supported by neither genetic nor morphological evidence. Meristic and morphometric variation appear insufficient to unambiguously and robustly diagnose troglobitic species and distinguish them from each other. Furthermore, beyond typical regressive troglomorphic traits, troglobitic species/ populations do not differ greatly in external morphology from their most closely related congener, the epigean species R. laticauda. Although statistically significant differences in body shape exist between troglobitic species/ populations and R. laticauda, such differences might not be the result of historical processes such as speciation but of convergent adaptation in phenotypically plastic traits instead. Based on the phylogenetic evidence presented here, continued recognition of all troglobitic species, as well as of the epigean R. nicaraguensis and R. parryi, implies a deep and generalized paraphyly in R. laticauda. Although the most parsimonious solution to this paraphyly is for the species involved to be synonymized with R. laticauda, we must admit that our geographic sampling of R. nicaraguensis and R. parryi is limited and based on vouchers unavailable for direct examination. Moreover, and more relevant to our research question, we recognize that the phylogenetic pattern uncovered herein is also consistent with a very recent divergence and early speciation of cave-dwelling populations from R. laticauda. Distinguishing between recent speciation of cave-dwelling lineages and intraspecific genetic structuring in R. laticauda (shaped by ongoing -albeit restricted- gene flow between epigean and hypogean populations), would therefore be the ultimate test for the validity of the current taxonomy. Discrimination of population and species boundaries in this system, however, will likely require additional comparative genetic data -ideally with genome-wide representation (e.g., genome-wide SNP data)- and state-of-the-art analytical approaches (e.g., coalescent-based species delimitation methods). Accordingly, in spite of the morphological and molecular evidence presented herein -which unarguably casts doubt on the validity of Mexican troglobitic species of Rhamdia- we refrain from making nomenclatural decisions until the abovementioned alternative evolutionary scenarios can be properly tested.











 nueva página del texto (beta)
nueva página del texto (beta)



