Introduction
The genus Amaranthus L. (Amaranthaceae Juss.) includes 70-75 monoecious and dioecious species with worldwide distribution, approximately half of which are native to the Americas (Costea et al., 2001; Das, 2016; Hernández-Ledesma et al., 2015; Iamonico, 2015).
Amaranthus is a critical genus from the taxonomic and nomenclatural point of view, exhibiting high degree of morphological diversity and a wide range of adaptability to different eco-geographical situations (Costea et al., 2001, 2003; Iamonico, 2014, 2015).
Amaranthus is economically very important, including cultivated plants (i.e., grain amaranths: A. caudatus, A. cruentus, and A. hypochondriacus) and potherbs (A. tricolor and A. blitum, many of which are also used as ornamentals (Pino et al., 2017; Sauer, 1967). Amaranthus also contains a suite of important agricultural and range weeds including: A. albus, A. hybridus, A. palmeri, A. powellii, A. retroflexus, A. spinosus, and A. tuberculatus (Costea, 2003; Pino et al., 2017; Sauer, 1967). Grain amaranths have received global attention, being a quality protein crop that is tolerant to abiotic stress (Akin-Idowu et al., 2016).
The origin of the grain amaranths is a subject of debate between 2 proposals: i) a single progenitor hypothesis, and ii) the independent domestication hypothesis (Sauer, 1967). No study has supported the independent domestication hypothesis (Kietlinski et al., 2014), while other studies have indicated that A. hybridus is the progenitor of grain amaranths, thereby supporting the single progenitor hypothesis (Kietlinski et al., 2014; Mallory et al., 2008; Maughan et al., 2011). Using phenotypic and genomic evidences, Stetter et al. (2017) suggested that some grain amaranths are incompletely domesticated species either because they were not strongly selected or had high levels of gene flow from their sympatric wild relatives that counteracted the fixation of key domestication traits found in the domesticated A. caudatus.
During the last 20 years, there has been an increased interest in understanding the diversity of Amaranthus at morphological, cytological, biochemical and molecular levels (Costea et al., 2006; Greizerstein et al., 1997; Iamonico 2010; Pratt et al., 2008; Stetter & Schmid, 2017). In particular, genetic data and phylogenetic relationships within and among crop species and their wild relatives are very important for the efficient utilization of plant genetic resources in breeding programs, determining phylogenetic relationships, developing ex situ conservation strategies of plant genetic resources and accurate identification of the plant taxa (Joshi, 2017; Marfila et al., 2015; Rao & Hodgkin, 2002).
RAPD analyses performed on grain amaranth (Adhikary & Pratt, 2015; Akin-Idowu et al., 2016; Popa et al., 2010; Sompornpailin & Khanthang, 2015; Stefunova et al., 2015), exhibited genetic diversity at intra and interspecific levels. At the interspecific level, Popa et al. (2010) found 37% polymorphism among grain amaranths collected from different geographic regions, whereas Akin-Idowu et al. (2016) revealed a higher rate, bordering at 81%. At the intraspecific level, the percentage of polymorphism varied between 70 and 100% (Stefunova et al., 2015) or from 19 to 72% (Sompornpailin & Khanthang, 2015). The percentage of polymorphism was found to be 71.43% in A. hypochondriacus, 100% in A. cruentus and A. caudatus, and 84.85% in A. tricolor (Patel et al., 2014). The cluster analysis of the RAPD data grouped accessions of A. hypochondriacus with those of both A. caudatus and A. cruentus (Sompornpailin & Khanthang, 2015), and revealed that A. cruentus stood apart while A. hypochondricus and A. caudatus overlapped (Mandal & Das, 2002).
The main aims of the present research were the following: 1) evaluate the genetic diversity among and within 25 accessions belonging to 10 wild and domesticated Amaranthus species, and 2) assess their phylogenetic relationships.
Materials and methods
Twenty-five accessions belonging to 10 different Amaranthus species were donated from the United States Department of Agriculture, Agricultural Research Service (USDA, ARS). The Amaranthus species represented were: A. hybridus, A. hypochondricus, A. palmeri, A. quitensis, A. retroflexus, A. spinosus, A. powellii, A. caudatus, A. cruentus, and A. tricolor (Table 1).
Table 1 Species name, plant name, accession number and origin of the germplasm studied.
|
No. |
Section |
Subsection |
Species |
Plant name |
Accession No. |
Origin |
|---|---|---|---|---|---|---|
|
1 |
Amaranthus |
Hybrida Mosyakin |
A. hybridus |
Ames 21188 |
PI 21188 |
South Africa |
|
2 |
& K. R. Robertson, |
RRC 847 |
PI 605351 |
Greece |
||
|
3 |
subsect. nova |
RRC 1195 |
PI 636181 |
USA ,Delaware |
||
| 4 |
GPAC 96-1 |
PI 23369 |
Brazil, Goias |
|||
|
5 |
Index seminum 110 |
PI 649304 |
Portugal, Coimbra |
|||
|
6 |
Hybrida Mosyakin |
A. hypochondriacus |
RRC 171 |
PI 274279 |
India, Himachal pradesh |
|
|
7 |
& K. R. Robertson, |
P373 |
PI 337611 |
Uganda |
||
|
8 |
subsect. nova |
RRC 1024 |
PI 477917 |
Mexico |
||
|
9 |
RRC 1004 |
PI 540446 |
Pakistan |
|||
|
10 |
Saueranthus |
A. palmeri |
Mapes 820 |
PI 604557 |
Mexico, Puebla |
|
|
11 |
Mosyakin & K. R. |
Pop 53 |
PI 607455 |
USA, Kansas |
||
|
12 |
Robertson, sect. nova |
Pop 59 |
PI 607461 |
USA, Kansas |
||
|
13 |
RRC 686 |
PI 632235 |
USA, Arizona |
|||
|
14 |
Amaranthus |
Amaranthus |
A. quitensis |
HH 70 |
PI 511744 |
Ecuador |
|
15 |
A. retroflexus |
DB, 8921 |
PI 572263 |
USA, lowa |
||
|
16 |
Centrusa Griseb. |
A. spinosus |
RRC 114 |
PI 619234 |
Indonesia, Sumatra |
|
|
17 |
Pyxidium Moquin in DC. Moquin in DC. Moquin in DC. |
A. tricolor |
PI 462129 |
India |
||
|
18 |
Amaranthus |
Hybrida Mosyakin & K. R. Robertson, subsect. nova |
A. powellii |
AMA 31/80 |
PI 572262 |
France |
|
19 |
Amaranthus |
Amaranthus |
A. caudatus |
Chua RRC 175 |
PI 166045 |
India |
|
20 |
RRC 279 |
PI 619264 |
Nepal |
|||
|
21 |
Love-Lies Bleeding |
PI 553073 |
USA, New Jersey |
|||
|
22 |
RRC 551 |
PI 511679 |
Argentina |
|||
|
23 |
Amaranthus |
Hybrida Mosyakin |
A. cruentus |
RRC 659 |
PI 451711 |
Mexico, Sonora |
|
24 |
& K. R. Robertson, |
RRC 685 |
PI 628793 |
Zaire, Shaba |
||
|
25 |
subsect. nova |
RRC384 |
PI 658727 |
Guatemala |
||
|
26 |
Amaranthus |
Hybrida Mosyakin |
A. hybridus |
Ames 21188 |
PI 21188 |
South Africa |
|
27 |
& K. R. Robertson, |
RRC 847 |
PI 605351 |
Greece |
||
|
28 |
subsect. nova |
RRC 1195 |
PI 636181 |
USA ,Delaware |
||
|
29 |
GPAC 96-1 |
PI 23369 |
Brazil, Goias |
|||
|
30 |
Index seminum 110 |
PI 649304 |
Portugal, Coimbra |
|||
|
31 |
Hybrida Mosyakin |
A. hypochondriacus |
RRC 171 |
PI 274279 |
India, Himachal pradesh |
|
|
32 |
& K. R. Robertson, |
P373 |
PI 337611 |
Uganda |
||
|
33 |
subsect. nova |
RRC 1024 |
PI 477917 |
Mexico |
||
|
34 |
RRC 1004 |
PI 540446 |
Pakistan |
|||
|
35 |
Saueranthus |
A. palmeri |
Mapes 820 |
PI 604557 |
Mexico, Puebla |
|
|
36 |
Mosyakin & K. R. |
Pop 53 |
PI 607455 |
USA, Kansas |
||
|
37 |
Robertson, sect. nova |
Pop 59 |
PI 607461 |
USA, Kansas |
||
|
38 |
RRC 686 |
PI 632235 |
USA, Arizona |
|||
|
39 |
Amaranthus |
Amaranthus |
A. quitensis |
HH 70 |
PI 511744 |
Ecuador |
|
40 |
A. retroflexus |
DB, 8921 |
PI 572263 |
USA, lowa |
||
|
41 |
Centrusa Griseb. |
A. spinosus |
RRC 114 |
PI 619234 |
Indonesia, Sumatra |
|
|
42 |
Pyxidium Moquin in DC. Moquin in DC. Moquin in DC. |
A. tricolor |
PI 462129 |
India |
||
|
43 |
Amaranthus |
Hybrida Mosyakin & K. R. Robertson, subsect. nova |
A. powellii |
AMA 31/80 |
PI 572262 |
France |
|
44 |
Amaranthus |
Amaranthus |
A. caudatus |
Chua RRC 175 |
PI 166045 |
India |
|
45 |
RRC 279 |
PI 619264 |
Nepal |
|||
|
46 |
Love-Lies Bleeding |
PI 553073 |
USA, New Jersey |
|||
|
47 |
RRC 551 |
PI 511679 |
Argentina |
|||
|
48 |
Amaranthus |
Hybrida Mosyakin |
A. cruentus |
RRC 659 |
PI 451711 |
Mexico, Sonora |
|
49 |
& K. R. Robertson, |
RRC 685 |
PI 628793 |
Zaire, Shaba |
||
|
50 |
subsect. nova |
RRC384 |
PI 658727 |
Guatemala |
||
Seeds were subjected to cold treatment for the first 24 h to improve germination, and were then germinated at 25-30 °C in media consisting of botmoss as a biological fertilizer mixed with silt in a 1:4 ratio, respectively. The terminal 4 leaves of plants at the 8-leaf stage were collected and stored in a -80 °C deep freezer until use for RAPD analysis. The stored leaf samples were then used for DNA isolation according to the CTAB method described by Doyle & Doyle (1990). Thus, 0.5 g of leaf stored samples were ground in liquid nitrogen, suspended in 1 ml preheated CTAB buffer (1.4 M NaCl, 0.2% β-mercaptoethanol, 100 mM Tris-Cl and 20 mM EDTA), and incubated at 65 °C for 1 h. The suspension was then centrifuged at 1,000 rpm. Later, 0.5 ml of a 24:1 v/ v chloroform: isomyl alcohol solution were added to the supernatant and centrifuged at 14,000 rpm. Ice-cold isopropanol was added to the aqueous layer to precipitate the nucleic acids (RNA and DNA), incubated at -20 °C overnight and centrifuged at 14,000 rpm. The pellet was washed carefully twice with cold 70% ethanol, dried at room temperature and re-dissolved in 100 µl of sterile deionized distilled water.
RAPD was performed as described by Williams et al. (1990) with minor modifications. Briefly, PCR amplification was performed in a 25 µl reaction mix containing 20-40 ng genomic DNA, 0.5 units of Taq polymerase (Sigma-Aldrich, St. Louis, MO, USA), 0.2 mM PCR Nucleotide Mix (Boehringer Mannheim, Tubingen, Germany), 0.6 µM RAPD primers (OPA-1 - OPA-4 and OPJ-11), 1 × reaction buffer IV (Advanced Biotechnologies Inc., Eldersburg, Maryland, USA), and 1.5 mM MgCl2. Amplification was performed for 45 cycles using a Biometera Uno thermal cycler (SPW Industrial, Laguna Hills, CA, USA), as follows: 1 cycle at 95 °C for 3 min and then 44 cycles at 92 °C for 2 min, 37 °C for 1 min and 72 °C for 2 min. The reactions were finally run at 72 °C for 10 min and further incubated on ice, at 4 °C. Five primers were selected for RAPD analysis based on their ability to amplify sections of the Amaranthus genome in reproducible amplification patterns. The names and sequences of the reproducible RAPD primers used in this study were the following: OPA-01 (5' CAGGCCCTTC 3'), OPA-02 (5' TGCCGAGCTG 3'), OPA-03 (5' AGTCAGCCAC 3'), OPA-04 (5' AATCGGGCTG 3'), and OPJ-11 (5' ACTCCTGCGA 3'). The amplification products were separated by electrophoresis on 2% agarose in 50× TAE buffer (Tris-Acetate EDTA buffer: 242 g Tris- base, 57.1 ml glacial acetic acid and 100 ml EDTA [0.5 M pH 8.0]), stained with 0.2 µg/ml ethidium bromide and photographed under UV light. The samples loaded were a combination of 10 µl PCR-product and 2 µl loading buffer. A 100-3,000 bp DNA ladder (Axygen, Union City, CA, USA) was used.
In RAPD analysis, the band identification was based on the mobility of DNA fragments and by numerous side-by-side comparisons of DNA extracts. The genetic diversity among the accessions was evaluated by the Jaccard similarity index and by multivariate analysis (cluster analysis and principal coordinate analysis). The analyses were performed using the frequencies of scored bands calculated for the accessions. The dendrogram was constructed through the average linkage-joining rule, using the “SYSTAT for Windows” software package, Version 7.0, 1997(SPSS Inc., San Jose, California, USA)
Results
Twenty primers were screened for the RAPD products. They were generated from DNA samples extracted from Amaranthus L. accessions. A total of 75 RAPD fragments were generated with 5 of 20 decamer arbitrary primers. The fragments generated by each primer showed variations in the total number, intensity, thickness and distance migrated down the agarose gel (Table 2; Fig. 1). Forty-seven fragments out of 75 were polymorphic, with 98.7% polymorphism. A fragment with a 450 bp molecular size, generated by OPA04, was the only monomorphic fragment in Amaranthus species (Fig. 1). The average number of fragments generated per primer ranged between 11 in OPA-01 and 17 in OPA-03 and OPA-04 with a total average number 7.1 (Table 3). The percentage of the polymorphism of the generated fragments by RAPD markers in the 10 species ranged between 100%, in A. quitensis, A. retroflexus, A. spinosus, A. powellii and A. tricolor, and 51.2%, in A. cruentus. Six fragments exhibited the lowest frequency (0.04) among the studied accessions (Table 2). These fragments can be considered accession-specific markers. They were the following: 600 and 300 bp fragments were unique markers for A. powellii (using the OPA-04 and OPJ-11 primers); a 350 bp fragment for A. tricolor PI 462129, from India (using the OPJ-11 primer); a 210 bp fragment for A. palmerii PI 607455, from Kansas (USA) (using the OPA-03 primer); a 500 bp fragment for A. caudatus PI 511679, from Argentina (using the OPA-04 primer), and a 700 bp fragment for A. quitensis PI 511744, from Ecuador (using the OPA-04 primer) (Fig. 2). These fragments can be considered as RAPD markers for these species.
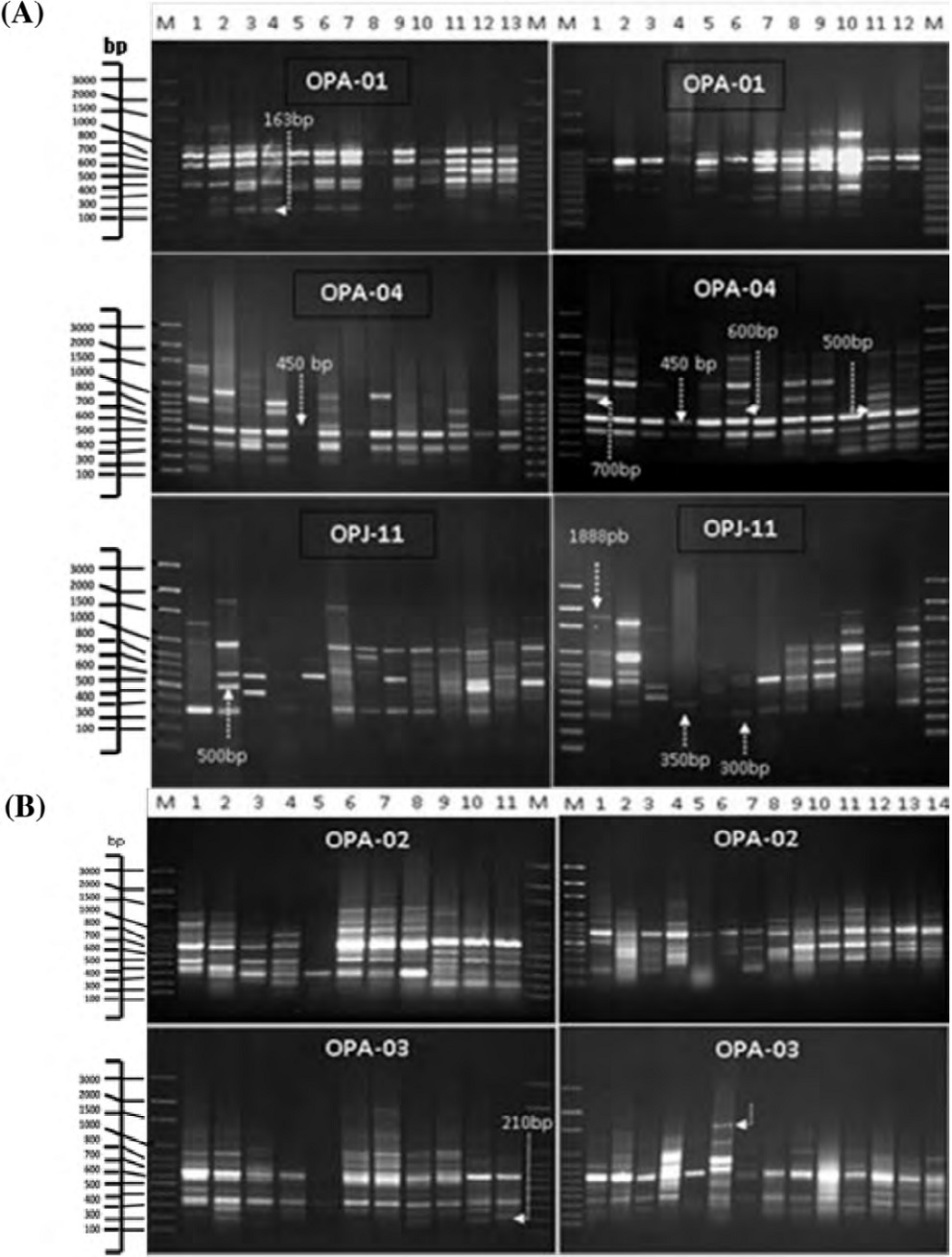
Figure 1 A, RAPD profiles show fragment amplification in the 25 different accessions of Amaranthus species that were analyzed using primers OPA-01, OPA-04 and OPJ-11. The profiles in each lane, including the DNA size markers (M) and the 25 accessions, were ordered following the same arrangement as that indicated in Table 1. B, RAPD profiles show fragment amplification in the 25 different accessions of Amaranthus species using primers OPA-02 and OPA-03. The profiles in each lane, including the DNA size markers (M) and the 25 accessions, were ordered following the same arrangement as that indicated in Table 1.
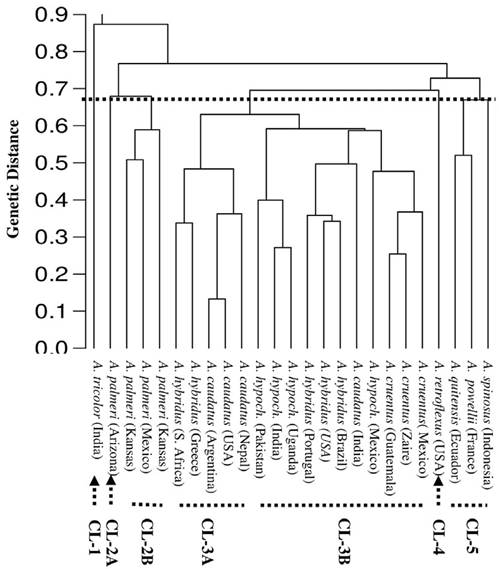
Figure 2 UPGMA dendrogram showing the relation among the 25 accessions of Amaranthus spp. generated by RAPD data.
Table 2 Survey of RAPD fragments detected in the 25 accessions of Amaranthus species analyzed using 5 random primers.
|
Primer |
Band number |
A. hybridus South Africa |
A. hybridus Greece |
A. hybridus USA Delaware |
A. hybridus Brazil |
A. hybridus Portugal |
A. hypo. India |
A. hypo. Uganda |
A. hypo. Mexico |
A. hypo. Pakistan |
A. palmeri Mexico |
A. palmeri USA Kansas |
A. palmeri USA Kansas |
A. palmeri USA Arizona |
A. quitensis Ecaudor |
A. retroflexus USA Iowa |
A. spinosus Indonesia |
A. tricolor India |
A. powellii France |
A. caudatus India |
A. caudatus Nepal |
A. caudatus New Jersey |
A. caudatus Argentina |
A. cruentus Mexico |
A. cruentus Zaire |
A. cruentus Guatemala |
Allele frequency |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
A1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
1 |
1 |
1 |
0 |
0 |
0.48 |
|
2 |
1 |
1 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
1 |
1 |
0 |
0 |
0.28 |
|
|
3 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
1 |
1 |
1 |
1 |
0 |
0 |
0.28 |
|
|
4 |
0 |
0 |
1 |
0 |
1 |
1 |
1 |
0 |
1 |
0 |
1 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0.36 |
|
|
5 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
0 |
1 |
1 |
1 |
1 |
1 |
0 |
1 |
1 |
1 |
1 |
0 |
1 |
1 |
1 |
1 |
1 |
0 |
0.84 |
|
|
6 |
0 |
0 |
1 |
0 |
0 |
1 |
1 |
0 |
0 |
1 |
1 |
1 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0.32 |
|
|
7 |
0 |
0 |
0 |
0 |
0 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0.12 |
|
|
8 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
0 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
0.96 |
|
|
9 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
0 |
0 |
0 |
0 |
1 |
1 |
1 |
0 |
0 |
0 |
1 |
0 |
1 |
1 |
1 |
1 |
1 |
1 |
0.68 |
|
|
10 |
1 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
1 |
1 |
1 |
0 |
0 |
0 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0.32 |
|
|
11 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
1 |
1 |
1 |
0 |
0 |
0.24 |
|
|
A2 |
12 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0.08 |
|
13 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
1 |
1 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0.28 |
|
|
14 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0.08 |
|
|
15 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
0 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
1 |
1 |
1 |
0.72 |
|
|
16 |
1 |
1 |
0 |
1 |
0 |
0 |
0 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
1 |
1 |
1 |
1 |
1 |
0.44 |
|
|
17 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
1 |
0 |
1 |
0 |
1 |
0 |
0 |
0 |
1 |
1 |
0.28 |
|
|
18 |
1 |
0 |
1 |
1 |
0 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
0 |
1 |
1 |
1 |
0 |
0 |
1 |
0 |
1 |
1 |
1 |
1 |
1 |
0.76 |
|
|
19 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0.08 |
|
|
20 |
0 |
0 |
0 |
0 |
0 |
1 |
1 |
0 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0.4 |
|
|
21 |
1 |
1 |
1 |
1 |
0 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
0.96 |
|
|
22 |
1 |
1 |
1 |
0 |
0 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
1 |
1 |
0 |
0 |
0 |
0.32 |
|
|
23 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0.12 |
|
|
24 |
1 |
1 |
1 |
1 |
0 |
1 |
1 |
1 |
1 |
0 |
0 |
0 |
1 |
1 |
0 |
1 |
0 |
1 |
0 |
1 |
1 |
1 |
1 |
1 |
1 |
0.72 |
|
|
25 |
1 |
1 |
0 |
0 |
0 |
1 |
1 |
0 |
1 |
0 |
0 |
0 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
1 |
1 |
1 |
0.44 |
|
|
26 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
1 |
0 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0.28 |
|
|
27 |
0 |
0 |
0 |
0 |
0 |
1 |
1 |
1 |
1 |
0 |
1 |
0 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0.28 |
|
|
A3 |
28 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
1 |
0.2 |
|
29 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
1 |
0 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0.24 |
|
|
30 |
0 |
1 |
1 |
1 |
1 |
٠ |
1 |
1 |
1 |
1 |
1 |
0 |
0 |
0 |
0 |
1 |
1 |
0 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
0.76 |
|
|
31 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
0 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
0.96 |
|
|
32 |
1 |
1 |
1 |
0 |
0 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
1 |
1 |
1 |
1 |
0.68 |
|
|
33 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
1 |
0 |
1 |
0 |
0 |
0 |
1 |
1 |
1 |
1 |
1 |
0.36 |
|
|
34 |
1 |
1 |
1 |
0 |
0 |
1 |
1 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
1 |
1 |
1 |
1 |
0.48 |
|
|
35 |
1 |
1 |
1 |
1 |
0 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
0 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
0.92 |
|
|
36 |
0 |
0 |
1 |
0 |
0 |
1 |
1 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0.24 |
|
|
37 |
1 |
1 |
0 |
0 |
0 |
1 |
1 |
1 |
1 |
0 |
0 |
1 |
1 |
0 |
1 |
0 |
1 |
0 |
0 |
1 |
1 |
1 |
0 |
0 |
0 |
0.52 |
|
|
38 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
1 |
1 |
0 |
0.2 |
|
|
39 |
1 |
1 |
1 |
0 |
0 |
1 |
1 |
1 |
1 |
0 |
0 |
0 |
1 |
0 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
1 |
0 |
0.48 |
|
|
40 |
0 |
0 |
1 |
0 |
0 |
1 |
1 |
1 |
1 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
1 |
1 |
0.4 |
|
|
41 |
1 |
1 |
0 |
0 |
0 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
1 |
1 |
1 |
0 |
0 |
0 |
0.36 |
|
|
42 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0.16 |
|
|
43 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
١ |
0 |
٠ |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0.04 |
|
|
44 |
0 |
0 |
0 |
0 |
0 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0.08 |
|
|
A4 |
45 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
1 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0.16 |
|
46 |
1 |
1 |
0 |
0 |
0 |
1 |
0 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
0 |
1 |
0 |
1 |
0 |
1 |
1 |
1 |
1 |
0 |
0 |
0.64 |
|
|
47 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
0 |
1 |
1 |
1 |
1 |
1 |
1 |
0 |
1 |
0.92 |
|
|
48 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0.12 |
|
|
49 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0.04 |
|
|
50 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
1 |
1 |
0 |
1 |
1 |
1 |
1 |
0 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
1 |
0.44 |
|
|
51 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0.04 |
|
|
52 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0.08 |
|
|
53 |
1 |
1 |
1 |
0 |
1 |
1 |
1 |
1 |
1 |
1 |
0 |
1 |
0 |
1 |
0 |
0 |
0 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
1 |
0.56 |
|
|
54 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
١ |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
٠ |
0.04 |
|
|
55 |
0 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
١ |
0 |
0 |
0 |
0 |
0 |
0 |
0.16 |
|
|
56 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0.04 |
|
|
57 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0.16 |
|
|
58 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
|
|
59 |
1 |
1 |
0 |
0 |
0 |
1 |
0 |
1 |
1 |
0 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0.28 |
|
|
60 |
1 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0.16 |
|
|
61 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0.12 |
|
|
Operon J11 |
62 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
0 |
1 |
1 |
0 |
1 |
1 |
0 |
0 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
0.84 |
|
63 |
0 |
0 |
0 |
0 |
0 |
1 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
1 |
0.28 |
|
|
64 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0.04 |
|
|
65 |
0 |
0 |
1 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
1 |
0 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0.24 |
|
|
66 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
1 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0.28 |
|
|
67 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
1 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0.16 |
|
|
68 |
0 |
1 |
1 |
1 |
1 |
0 |
0 |
1 |
1 |
0 |
0 |
1 |
0 |
1 |
1 |
0 |
0 |
0 |
1 |
1 |
1 |
1 |
0 |
0 |
0 |
0.52 |
|
|
69 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
1 |
1 |
0 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
1 |
1 |
1 |
0.36 |
|
|
70 |
0 |
0 |
1 |
1 |
1 |
1 |
1 |
0 |
1 |
0 |
1 |
0 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0.4 |
|
|
71 |
1 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
1 |
0 |
1 |
1 |
0.32 |
|
|
72 |
1 |
1 |
0 |
1 |
0 |
1 |
1 |
1 |
1 |
1 |
1 |
0 |
1 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
1 |
1 |
1 |
1 |
0.68 |
|
|
73 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0.04 |
|
|
74 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0.16 |
|
|
75 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0.04 |
|
|
No. of alleles |
38 |
38 |
27 |
21 |
14 |
40 |
35 |
28 |
37 |
26 |
35 |
28 |
35 |
23 |
24 |
20 |
14 |
22 |
16 |
20 |
28 |
30 |
35 |
25 |
27 |
Table 3 The total number of amplified RAPD fragments generated, and number of polymorphic and monomorphic fragments produced by each primer.
| Species name | Total number of generated fragments | No. of monomorphic fragments | No. of polymorphic fragments | Percent of polymorphism | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 | P2 | P3 | P4 | P5 | P1 | P2 | P3 | P4 | P5 | P1 | P2 | P3 | P4 | P5 | ||
| A. hybridus | 10 | 10 | 14 | 10 | 9 | 5 | 1 | 1 | 2 | 1 | 5 | 9 | 10 | 8 | 8 | 74.5 |
| A. hypochondriacus | 9 | 11 | 13 | 10 | 11 | 2 | 5 | 9 | 3 | 2 | 7 | 6 | 4 | 7 | 9 | 61.1 |
| A. palmeri | 8 | 10 | 14 | 13 | 12 | 4 | 4 | 3 | 4 | 0 | 4 | 6 | 11 | 9 | 12 | 73.6 |
| A. quitensis | 3 | 8 | 3 | 5 | 5 | - | - | - | - | - | 3 | 8 | 3 | 5 | 5 | 100 |
| A. retroflexus | 3 | 6 | 7 | 6 | 6 | - | - | - | - | - | 3 | 6 | 7 | 6 | 6 | 100 |
| A. spinosus | 3 | 8 | 3 | 5 | 3 | - | - | - | - | - | 3 | 8 | 3 | 5 | 3 | 100 |
| A. tricolor | 4 | 2 | 7 | 1 | 1 | - | - | - | - | - | 4 | 2 | 7 | 1 | 1 | 100 |
| A. powellii | 7 | 4 | 4 | 6 | 2 | - | - | - | - | - | 7 | 4 | 4 | 6 | 2 | 100 |
| A. caudatus | 8 | 11 | 8 | 4 | 7 | 3 | 1 | 3 | 2 | 2 | 5 | 10 | 5 | 2 | 5 | 71.1 |
| A. cruentus | 9 | 7 | 11 | 6 | 8 | 3 | 6 | 7 | 1 | 3 | 6 | 1 | 4 | 5 | 5 | 51.2 |
| Average | 6.4 | 7.7 | 8.4 | 6.6 | 6.4 | 3.4 | 3.4 | 4.6 | 2.4 | 1.6 | 4.7 | 6 | 5.8 | 5.4 | 5.6 | |
| Total average | 7.1 | 3.08 | 5.5 | 83.2 | ||||||||||||
The level of genetic diversity among RAPD fragments was calculated with the Jaccard coefficient of similarity (Table 4). The overall mean similarity index calculated by Jaccard similarity index (JSI) for pair wise combinations of the amplified fragments generated by the 5 arbitrary primers on the genomic DNA of the 25 accessions of Amaranthus species ranged from 0.11 to 0.880 with an average of 0.430. The highest similarity index (0.880) was between A. caudatus PI 553073, from New Jersey (USA) and A. caudatus PI 511679, from Argentina. Two higher values 0.714 and 0.766 for similarity indices were detected between A. cruentus PI 451711, from Sonora (Mexico) and A. cruentus PI 628793, from Shaba (Zaire). The lowest index (0.11) was observed between A. tricolor PI 462129, from India, and A. palmeri PI 604557, from Puebla (Mexico).
The matrix of eigenvectors and values of the principle components (PCs) resulting from the interaction of the RAPD data (Table 5) indicated that all the DNA fragments generated by the 5 primers influenced 56.72% of the variability accumulated by the first 4 components of the PCA. The first component explained 34.85% of the total diversity, while the second to fourth components explained 8.29%, 6.96% and 6.62% of the total diversity, respectively. All accessions were separated on the first component except the accession PI 607455 of A. palmeri, from Kansas (USA) and the accession PI 632235 of A. palmeri, from Arizona (USA) were separated on the second and fourth components respectively. Also, the accession PI 1619234 of A. spinosus, from Indonesia, was separated on the third component.
The phenogram constructed using each accession as an Operational Taxonomic Unit and including all the DNA fragments generated by the 5 primers is presented in figure 2. At a genetic distance of 68%, the accessions were divided genetically into 5 clusters. The first cluster separated A. tricolor from India in a distinct branch away from the rest of the accessions. The second cluster gathered all accessions of A. palmeri and was divided into 2 sub clusters (2A and 2B). The A. palmeri, from Arizona (USA), was separated into sub cluster 2A, while the other 3 accessions of A. palmeri from, Kansas (USA) and Mexico, were separated in sub cluster 2B. The third cluster contained more than 50% of the accessions studied and was subdivided into 2 sub clusters (3A and 3B). The sub cluster 3A included 2 accessions of A. hybridus, from South Africa and Greece, and 3 accessions of A. caudatus, from Argentina, USA and Nepal. The sub cluster 3B contained 3 accessions of A. hypochondriacus, from Pakistan, India and Uganda, 3 accessions of A. hybridus, from Portugal, USA and Brazil, and 3 accessions of A. cruentus, from Guatemala, Zaire and Mexico. The fourth cluster was comprised only of A. retroflexus. This accession is genetically distinct from other accessions, being separated at a genetic distance of 70%. The fifth cluster included A. quitensis, from Ecuador, A. powellii, from France and A. spinosus, from Indonesia.
Discussion
Genetic diversity among 25 accessions belonging to the 10 Amaranthus species studied was assessed with 75 RAPD polymorphic fragments. The RAPD polymorphic fragments were different in number (11 to 17), intensity and position. The diversity between the generated fragments depends on primers used, DNA sequence, number of accessions and the extent of diversity in these accessions (Akin-Idowu et al., 2016; Popa et al., 2010; Stefunova et al., 2015). The percentage of polymorphism between the accessions studied was 98.7. This percentage was higher than that reported by Ray and Roy (2008), Popa et al. (2010) and Lymanskaya (2012) which ranged between 37 and 85%. The discrepancy between data here reported and the results described by these 3 groups could be attributed to 2 main factors. First, these studies were performed on grain amaranth, which has a relative narrow genetic diversity due to the selection pressures of domestication, and second, the primers used in one of the studies amplified mostly the conserved part of the genome, being therefore unable to detect any significant variation within a population (Popa et al., 2010).
The low value of RAPD polymorphism found in grain Amaranthus species compared to the wild Amaranthus species reflects a narrow range of intra-specific diversity in the grain species. This pattern of genetic diversity in grain Amaranthus spp. suggests that they may have passed through genetic bottlenecks during the process of speciation and/or experienced strong directional selection as a result of domestication.
The cultivated A. tricolor PI 462129 and A. hybridus PI 649304 were characterized by having the lowest number of RAPD fragments (14 fragments), whereas the most polymorphic species was A. hypochondriacus PI 274279 (40 fragments). This finding agreed with the work of Ray and Roy (2008).
The output of the Jaccard binary similarity coefficient and cluster analysis based on all DNA fragments generated by the 5 primers showed that the strongest homogeneity was found between A. caudatus PI 553073, from New Jersey (USA), A. caudatus PI 619264, from Nepal, A. hypochondriacus PI 337611, from Uganda, A. hypochondriacus PI 274279, from India, A. cruentus PI 628793, from Zaire and A. cruentus PI 451711, from Sonora (Mexico). This homogeneity might be attributed to the cultivated nature of these species. The Jaccard similarity coefficient data showed that A. hypochondriacus (0.461) was the grain amaranth closest to A. hybridus, followed by A. caudatus (0.436) and A. cruentus (0.349). This sequence of evolution was different from that suggested by the single progenitor hypothesis postulated by Sauer (1967). The similarity coefficient between A. tricolor and other species ranged between 0.011 and 0.185, showing that A. tricolor is genetically distant from other amaranth species. This finding agreed with data reported by Patel et al. (2014), who indicated that A. tricolor had the greatest genetic distance from grain amaranth.
Tabla 4 Within accession genetic diversity estimated as Jaccard binary similarity coefficients (SJC) calculated from pairwise combination of the amplified fragments generated by the 5 arbitrary primers on the genomic DNA of the 25 accessions of Amaranthus species used in this study.
|
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
15 |
16 |
17 |
18 |
19 |
20 |
21 |
22 |
23 |
24 |
25 |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
1 |
1.000 |
||||||||||||||||||||||||
|
2 |
0.680 |
1.000 |
|||||||||||||||||||||||
|
3 |
0.296 |
0.351 |
1.000 |
||||||||||||||||||||||
|
4 |
0.378 |
0.378 |
0.646 |
1.000 |
|||||||||||||||||||||
|
5 |
0.199 |
0.336 |
0.639 |
0.616 |
1.000 |
||||||||||||||||||||
|
6 |
0.360 |
0.253 |
0.479 |
0.226 |
0.311 |
1.000 |
|||||||||||||||||||
|
7 |
0.335 |
0.335 |
0.635 |
0.369 |
0.444 |
0.714 |
1.000 |
||||||||||||||||||
|
8 |
0.265 |
0.376 |
0.455 |
0.378 |
0.267 |
0.390 |
0.383 |
1.000 |
|||||||||||||||||
|
9 |
0.174 |
0.280 |
0.538 |
0.394 |
0.417 |
0.656 |
0.520 |
0.562 |
1.000 |
||||||||||||||||
|
10 |
0.158 |
0.214 |
0.271 |
0.357 |
0.226 |
0.176 |
0.217 |
0.365 |
0.346 |
1.000 |
|||||||||||||||
|
11 |
0.121 |
0.014 |
0.189 |
0.310 |
0.238 |
0.071 |
0.196 |
0.162 |
0.306 |
0.498 |
1.000 |
||||||||||||||
|
12 |
0.321 |
0.210 |
0.340 |
0.317 |
0.408 |
0.335 |
0.273 |
0.316 |
0.286 |
0.422 |
0.383 |
1.000 |
|||||||||||||
|
13 |
0.282 |
0.175 |
0.189 |
0.250 |
0.169 |
0.339 |
0.304 |
0.162 |
0.360 |
0.273 |
0.464 |
0.328 |
1.000 |
||||||||||||
|
14 |
0.194 |
0.136 |
0.224 |
0.358 |
0.275 |
0.274 |
0.247 |
0.383 |
0.327 |
0.184 |
0.131 |
0.383 |
0.131 |
1.000 |
|||||||||||
|
15 |
0.162 |
0.162 |
0.379 |
0.463 |
0.332 |
0.355 |
0.332 |
0.416 |
0.467 |
0.401 |
0.218 |
0.475 |
0.218 |
0.226 |
1.000 |
||||||||||
|
16 |
0.113 |
0.113 |
0.176 |
0.363 |
0.098 |
0.020 |
0.040 |
0.345 |
0.249 |
0.321 |
0.342 |
0.158 |
0.222 |
0.318 |
0.103 |
1.000 |
|||||||||
|
17 |
0.062 |
0.131 |
0.068 |
0.082 |
0.122 |
0.037 |
0.238 |
0.125 |
0.075 |
0.011 |
0.037 |
0.125 |
0.101 |
0.052 |
0.185 |
0.021 |
1.000 |
||||||||
|
18 |
0.226 |
0.284 |
0.249 |
0.250 |
0.293 |
0.250 |
0.278 |
0.229 |
0.184 |
0.331 |
0.337 |
0.411 |
0.278 |
0.461 |
0.123 |
0.340 |
0.067 |
1.000 |
|||||||
|
19 |
0.058 |
0.188 |
0.355 |
0.473 |
0.419 |
0.161 |
0.165 |
0.338 |
0.332 |
0.305 |
0.231 |
0.338 |
0.035 |
0.360 |
0.480 |
0.201 |
0.085 |
0.165 |
1.000 |
||||||
|
20 |
0.354 |
0.474 |
0.490 |
0.497 |
0.562 |
0.322 |
0.403 |
0.407 |
0.310 |
0.194 |
0.101 |
0.345 |
0.222 |
0.449 |
0.233 |
0.318 |
0.175 |
0.539 |
0.422 |
1.000 |
|||||
|
21 |
0.486 |
0.596 |
0.455 |
0.562 |
0.338 |
0.225 |
0.383 |
0.430 |
0.286 |
0.249 |
0.162 |
0.259 |
0.107 |
0.264 |
0.298 |
0.283 |
0.125 |
0.350 |
0.338 |
0.657 |
1.000 |
||||
|
22 |
0.588 |
0.588 |
0.408 |
0.521 |
0.307 |
0.273 |
0.382 |
0.383 |
0.229 |
0.206 |
0.109 |
0.214 |
0.164 |
0.283 |
0.257 |
0.246 |
0.098 |
0.251 |
0.306 |
0.615 |
0.889 |
1.000 |
|||
|
23 |
0.335 |
0.228 |
0.412 |
0.429 |
0.306 |
0.286 |
0.304 |
0.383 |
0.360 |
0.273 |
0.250 |
0.273 |
0.089 |
0.305 |
0.275 |
0.222 |
0.101 |
0.278 |
0.296 |
0.282 |
0.494 |
0.491 |
1.000 |
||
|
24 |
0.302 |
0.358 |
0.412 |
0.441 |
0.242 |
0.265 |
0.416 |
0.565 |
0.321 |
0.436 |
0.132 |
0.214 |
0.076 |
0.266 |
0.424 |
0.213 |
0.169 |
0.228 |
0.322 |
0.277 |
0.448 |
0.462 |
0.586 |
1.000 |
|
|
25 |
0.240 |
0.296 |
0.421 |
0.460 |
0.354 |
0.367 |
0.468 |
0.627 |
0.427 |
0.388 |
0.134 |
0.283 |
0.078 |
0.465 |
0.379 |
0.239 |
0.068 |
0.249 |
0.355 |
0.302 |
0.397 |
0.408 |
0.523 |
0.766 |
1.000 |
Cluster analysis based on the RAPD data reported in the present study was more reliable, considering that most of the accessions belonging to the same species were collected together. Conversely, accessions from different geographical origins were relatively unique and tended to cluster in specific sections of the dendrogram. This information suggested that the diversity detected was determined not only by environmental differences but also by genetic factors (Govindaraj et al., 2015).
The edible A. tricolor appeared in a distinct branch, apart from all other studied species. This finding agreed with the findings of Xu and Sun (2001) and Mosyakin and Robertson (1996), who accepted the inclusion of A. tricolor into a different subgenus, Albersia (Kunth) Gren. & Godr., that was able to accommodate this species within those having indehiscent utricles. The accessions of A. palmeri clustered into a distinct group that was separated from the other species, thereby supporting its classification into a distinct subgenus, Acnida (L.) Aellen ex K.R. Robertson sensuMosyakin and Robertson (1996). The A. cruentus and A. palmeri species formed independent taxonomic units with numerous loci that allowed them to be separated from each other and from all other species. A. hybridus accessions clustered together with grain amaranth, a result that contrary to other results derived from the present study (see above), confirmed the single progenitor hypothesis for grain amaranth suggested by Sauer (1967).
Table 5 Matrix of eigenvectors and values of the principal components analysis (PCA) based on RAPD data for the accessions of Amaranthus species analyzed.
| Species | Principal components | |||
|---|---|---|---|---|
| C1 | C2 | C3 | C4 | |
| A. hybridus | 0.533 | -0.426 | 0.103 | 0.351 |
| A. hybridus | 0.575 | -0.506 | 0.078 | 0.230 |
| A. hybridus | 0.722 | -0.008 | -0.281 | 0.082 |
| A. hybridus | 0.741 | -0.025 | 0.113 | -0.092 |
| A. hybridus | 0.629 | 0.059 | -0.101 | 0.133 |
| A. hypochondriacus | 0.570 | 0.111 | -0.526 | 0.358 |
| A. hypochondriacus | 0.656 | -0.004 | -0.464 | 0.291 |
| A. hypochondriacus | 0.682 | 0.045 | -0.143 | -0.267 |
| A. hypochondriacus | 0.651 | 0.316 | -0.361 | 0.090 |
| A. palmeri | 0.522 | 0.417 | 0.222 | -0.146 |
| A. palmeri | 0.380 | 0.559 | 0.383 | 0.159 |
| A. palmeri | 0.560 | 0.345 | 0.135 | 0.187 |
| A. palmeri | 0.374 | 0.360 | 0.121 | 0.553 |
| A. quitensis | 0.526 | 0.127 | 0.176 | -0.127 |
| A. retroflexus | 0.575 | 0.296 | -0.224 | -0.170 |
| A. spinosus | 0.394 | 0.193 | 0.520 | -0.147 |
| A. tricolor | 0.185 | -0.079 | -0.132 | 0.044 |
| A. powellii | 0.507 | 0.125 | 0.424 | 0.232 |
| A. caudatus | 0.537 | 0.159 | 0.069 | -0.331 |
| A. caudatus | 0.696 | -0.251 | 0.227 | 0.179 |
| A. caudatus | 0.719 | -0.475 | 0.232 | -0.030 |
| A. caudatus | 0.692 | -0.537 | 0.192 | 0.005 |
| A. cruentus | 0.618 | -0.088 | 0.010 | -0.289 |
| A. cruentus | 0.661 | -0.096 | -0.136 | -0.469 |
| A. cruentus | 0.690 | 0.013 | -0.191 | -0.444 |
| Variance explained by components | 8.712 | 2.073 | 1.740 | 1.654 |
| Percent of total variance explained | 34.846 | 8.291 | 6.962 | 6.616 |
| Accumulated eigenvectors | 34.846 | 43.137 | 50.99 | 56.715 |
The first 4 components of the PCA based on all DNA fragments generated by the 5 primers showed a total diversity of 56.72% and the separation of most of the accessions analyzed on component 1, indicating a high degree of correlation among all accessions. A. spinosus was separated into a distinct third component indicating the low similarity existing between this and other amaranth species, in accordance to its classification in a distinct section: Centrusa Griseb. sensuMosyakin & Robertson (1996). A. palmeri PI 607455, from Kansas (USA), and A. palmeri PI 632235, from Arizona (USA), were also separated on the second and fourth components, respectively. The separation of A. palmeri accessions (an important herbicide-resistant weed species) on different axes agreed with the recent finding of Waselkov et al. (2018) who analyzed the phylogeny of the genus Amaranthus based on several low-copy nuclear loci and chloroplast regions. The high genetic variability observed in A. palmeri population could be attributed to: 1) the wide distribution geographical range of A. palmeri, which extends from northwestern Mexico and southern California, to New Mexico and Texas (Sauer 1957), and 2) A. palmeri is an obligate out crosser that is wind pollinated, a property that greatly favors a genetically variable population (Ward et al., 2013).











 nova página do texto(beta)
nova página do texto(beta)


