Introduction
On May 16, 1898, E. W. Nelson and E. A. Goldman collected 5 adult and 1 immature jackrabbits on the coastal plain about 10 miles north of Altamira in southern Tamaulipas (Nelson, 1909). Goldman (1951) described the habitat at that time as mostly “chaparral” (= thornscrub), characterized by open grassy plains with patches of guayaba bushes (Psidium guajava), mesquites (Prosopis juliflora), acacias, and cactuses of various species scattered over the plains.
The type specimen for the series (not a member of the white-sided or black-tailed groups), which Nelson (1904) initially described as the Alta Mira jackrabbit (Lepus merriami altamirae), was said to be restricted to the coastal plain of southern Tamaulipas, extreme northern Veracruz and the eastern border of San Luis Potosí, Mexico. L. merriami had been described by Mearns in 1896 as a new species very similar to Lepus callotis of Mexico but with shorter ears with black tips instead of white (Mearns, 1896). Nelson (1904) noted that the Alta Mira jackrabbit had a limited distribution, occupied the southernmost area of the species range along the Gulf Coast of Mexico, and probably did not occur as far north as Victoria, Tamaulipas. Nelson (1909), later described the animal’s elevational range as between sea level and 150 m within the “Arid Tropical” zonal range.
In a later monograph on the rabbits of North America, Nelson (1909) revised his taxonomic assignation, elevating the Tamaulipas jackrabbit to the level of species, and noted that more careful examination showed its relationship to be with the white-sided group of jackrabbits and most alike in general appearance to L. callotis (white-sided jackrabbit), a similarity also noted by Mearns in his original description of L. merriami (Mearns, 1896). Although the color of the upperparts resembled that of L. merriami (currently considered a subspecies of Lepus californicus, black-tailed jackrabbit, distributed in south Texas and northern Tamaulipas), Nelson (1909) considered the absence of a black patch on the posterior half of the ear at the tip and the white flanks (somewhat obscured in some of the original specimens) to be strong characteristics, which place L. altamirae in the callotis group. Nelson also cautioned that L. altamirae, although a “well-marked species,” was less strongly differentiated from L. californicus (as “L. merriami”) than L. callotis and L. flavigularis (Tehuantepec jackrabbit), owing mainly to the less definite segregation of white on the sides.
Nelson (1909), who had observed L. altamirae in the field, described other characters of L. altamirae that were suggestive of a relationship with L. callotis and the white-sided group of jackrabbits. These included an iron gray rump patch and a nape with 2 lateral black bands (Fig. 1) extending back from the base of the ears separated by a buffy median. The underside of the neck was described as “deeper and clearer buffy” with the “posterior half of ears white without any trace of black at tip; white of underparts extending up on flanks nearly as high as in L. callotis”. The skull of L. altamirae was said to resemble that of L. callotis in general, but “much larger and proportionally narrower”. The average measurements of 5 adults, all collected by Nelson and Goldman, were: length: 587 mm, tail: 72 mm, hind foot: 136 mm, ear: 110.6 mm. Nelson’s distribution map of white-sided jackrabbits (Nelson, 1909) places L. altamirae as an isolated population in extreme southern Tamaulipas, south of the Sierra de Tamaulipas, where no black-tailed jackrabbits occur (Fig. 2).
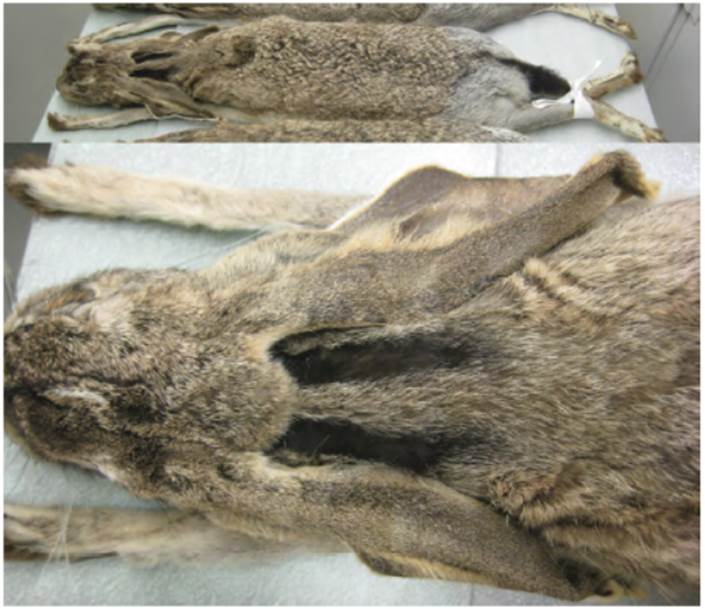
Figure 1 Dorsal and lateral views of a museum specimen of L. altamirae (USNM 93692) showing black lines on nape, lack of black ear tips and other characteristics of L. callotis and white-sided jackrabbits.

Figure 2 Geographical distribution of L. californicus merriami, L. californicus curti, and L. altamirae.
More than 40 years later, in March 1950, E. R. Hall and C. von Wedel visited Boca Jesús María, on Tamaulipas’ barrier beach 144 km south and 16 km west of the Texas border at Matamoros (24°34’ N, 97°39’ W, University of Kansas Biodiversity Institute. KUBI Mammalogy Collection, http://portal.vertnet.org/o/ku/kum?id=91870c9d-1ed8-11e3-bfac-90b11c41863e), which is over 100 km north of Nelson and Goldman’s 1898 collection site. Here, Hall and von Wedel collected 4 adult female jackrabbits in worn pelage, which Hall (1951a) described as Lepus californicus curti. This taxon was described as paler, smaller and with shorter ears than Lepus californicus merriami of southern Texas. On comparing it with Nelson’s L. altamirae specimens, he questioned the latter’s inclusion in the L. callotis group as he noted that 3 of the specimens from north of Altamira had some dusky coloration on the ears and otherwise exhibited pelage characters that he considered similar to his L. c. curti. Hall considered L. c. curti to be intermediate between L. c. merriami and L. altamirae and placed all 3 in the L. californicus or black-tailed group of jackrabbits. His basis for including what he called L. californicus altamirae in the L. californicus group was a longer palate, which he considered more characteristic of black-tailed jackrabbits than the L. callotis group. Nonetheless, he noted that skin measurements indicated that L. altamirae was significantly larger than L. c. curti (e.g., the hind foot measurements of L. altamirae ranged from 136 to 142 mm vs. 115 to 125 for L. c. curti).
Although Hall never saw a live specimen of L. altamirae, he continued to place it in the black-tailed group in his revision of North American lagomorphs (Hall, 1951b). He did so despite having given up on using skull characters to define the 2 groups, noting that: “A certain means for distinguishing the skulls of the black-tailed jackrabbit from those of all of the white-sided rabbits has not yet been found” (Hall 1951b). In addition, Hall stated that, in Tamaulipas (where, he affirmed, only black-tailed jackrabbits occur), this taxon presented extensively white flanks, and that some individuals lacked the terminal black patch on the ear.
Álvarez (1963), in his treatment of the mammals of Tamaulipas examined only 2 jackrabbit specimens, both from north of Soto La Marina -a locality east of Ciudad Victoria and north of the Sierra de Tamaulipas, approximately 200 km from Altamira, where neither Nelson nor Hall had collected jackrabbits (Fig. 2). Following Hall’s designation, he identified both animals as L. c. altamirae.
In view of the conflicting taxonomic assignations, it is uncertain what the status of the Tamaulipas jackrabbit is. Was Nelson correct in placing L. altamirae in the white-sided group and describing it as a separate species of white-sided jackrabbit? Or were the specimens he labeled L. altamirae a subspecies of black-tailed jackrabbit as determined by Hall (1981)? A reading of Nelson’s habitat descriptions and assignment of L. altamirae to a tropic-subtropic distribution also suggested that this taxon belonged to the white-sided jackrabbit group.
To resolve the question as to which group -white-sided or black-tailed- for the specimens that originated from northwest of Altamira, Tamaulipas, we assessed the pelage characteristics of the 5 specimens of L. altamirae collected by E. W. Nelson in 1898 currently cataloged at the National Museum. In addition, we performed phylogenetic analyses using the mitochondrial cytochrome b gene (MT-CYB) to determine if L. altamirae is most closely related to members of the white-sided or black-tailed group of jackrabbits.
Materials and methods
We examined the pelage characteristics of the 5 specimens of L. altamirae at the National Museum (USNM: 92981, 92982, 93692, 93693, 93694), currently identified as Lepus californicus melanotis (Texas black-tailed jackrabbit) and compared them to specimens of L. callotis and other white-sided jackrabbits.
For DNA analyses, we collected scrapings of dried skin tissue from 2 adult female specimens of L. altamirae (USNM 93692 and USNM 93694). We initially pulverized the skin tissue samples with stainless steel beating beads (Next Advance, Inc. Averill Park, New York) and a high-speed “beater” (Benchmark Scientific, Inc. Edison, New Jersey). We added 400 µl of lysis buffer (50 mM tris pH 8.0, 50 mM EDTA, 25 mM sucrose, 100 mM NaCl, 1.0% SDS, 40 mM DTT) and 25 µl of 20 mg/ml proteinase K to the powdered samples and incubated them for 48 hours at 55 °C with agitation in 2.0 ml microcentrifuge tubes. We followed a phenol: chloroform: isoamylalcohol (25:24:1) extraction protocol utilizing light and heavy phase lock gel tubes (5 PRIME, Gaithersburg, Maryland) to isolate DNA from the lysate, and we added 3M NaOAc and isopropanol to precipitate the DNA. We performed ethanol washes to further purify the DNA before re-suspending it in 60 µl of low TE, pH 8.0.
We designed 5 sets of primers (Table 1) using Primer-BLAST (Ye et al., 2012) to amplify small regions of the mitochondrial cytochrome b gene (MT-CYB), appropriate for degraded DNA from museum specimens. We performed PCR amplifications in a total reaction volume of 20 µl containing 1X PCR buffer (Invitrogen™, Thermo Fisher Scientific Inc. Waltham, Massachusetts), 1.5 mM MgCl2, 0.2 mM dNTPs, 0.05% BSA (Sigma-Aldrich, St. Louis, Missouri), 1U of Taq DNA Polymerase (Invitrogen™), 0.5 μM of each forward and reverse primers, and 15 ng of template DNA. PCR conditions consisted of initial denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 63 °C for 1 min, extension at 72 °C for 1 min, and a final extension at 72 °C for 10 min. We used a Mastercycler PCR machine (Eppendorf, Westbury, New York) for PCR. We sent resultant MT-CYB PCR products to the University of Arizona Genetics Core facility to be sequenced in forward and reverse directions on an ABI 3100 automated DNA analyzer (Applied Biosystems Inc., Foster City, California).
Table 1 Set of primers designed in Primer-BLAST (Ye et al., 2012) to amplify short fragments of MT-CYB. bp = base pairs.
| Primer | Sequence | Fragment length |
| Lepus240_F1 | 5'-ttacctgcaygctaacggag-3’ | |
| Lepus381_R1 | 5'-tgctgtggctattactgcga-3’ | 142 bp |
| Lepus526_F2 | 5'-acccgattcttcgctttcca-3’ | |
| Lepus627_R2 | 5'-tgaagggttattggagcccg-3’ | 102 bp |
| Lepus753_F3 | 5'-cgacccagacaactacaccc-3’ | |
| Lepus863_R3 | 5'-agtttgttggggatrgagcg-3’ | 111 bp |
| Lepus829_F4 | 5'-gcctaygccattttacgctcy-3’ | |
| Lepus967_R4 | 5'-cttggctgatgggtcggaat-3’ | 139 bp |
| Lepus948_F5 | 5'-attccgacccatcagccaag-3’ | |
| Lepus1037_R5 | 5'-gggtgttcractggytgtcct-3’ | 90 bp |
To examine interspecific relationships of L. altamirae specimens, we compared sequences from the 2 museum samples to MT-CYB sequences (613 to 1,140 bp) of 11 representatives of white-sided jackrabbits (L. callotis, L. flavigularis, and L. alleni), 7 black-tailed jackrabbits (L. californicus), and 3 white-tailed jackrabbits (Lepus townsendii) obtained from GenBank. We used sequences of the genera Sylvilagus and Romerolagus as outgroups. We downloaded these sequences from the publicly available GenBank database (http://www.ncbi.nlm.nih.gov; Table 2).
Table 2 Species used for phylogenetic reconstruction, localities, GenBank accession numbers, length of the fragment in base pairs (bp), and source. Group: white-sided jackrabbits (WS), black-tailed jackrabbits (BT), white-tailed jackrabbits (WT), outgroup (OG). Sources: a, Ramírez-Silva et al. (2010); b, Halanych et al. (1999)); , Lorenzo et al. (2014); d, Halanych & Robinson (1999); e, Matthee et al. (2004); f, C. Lorenzo, personal communication, August 19, 2016; ‘---’ indicates no available information.
| Group | Species | Locality | GenBank number | bp | Source |
| WS | Lepus alleni | Sonora, Mexico | HQ596458 | 1,140 | a |
| WS | Lepus alleni | Navojoa, Sonora, Mexico | AF010156 | 702 | b |
| WS | Lepus alleni | Tiburón Island, Mexico | AF010157 | 702 | b |
| WS | Lepus callotis | Jalisco, Mexico | HQ596469 | 1,140 | a |
| WS | Lepus callotis | Chihuahua, Mexico | HQ596467 | 1,140 | a |
| WS | Lepus callotis | Hidalgo county, New Mexico, USA | AF010159 | 702 | b |
| WS | Lepus callotis | Hidalgo county, New Mexico, USA | AF010158 | 702 | b |
| WS | Lepus callotis | Jalisco, Mexico | HQ596468 | 1,140 | a |
| WS | Lepus flavigularis | Oaxaca, Mexico | HQ596475 | 1,140 | a |
| WS | Lepus flavigularis | Oaxaca, Mexico | KT308125 | 1,140 | c |
| WS | Lepus flavigularis | Oaxaca, Mexico | --- | 613 | f |
| BT | Lepus californicus | Chihuahua, Mexico | HQ596462 | 1,140 | a |
| BT | Lepus californicus | Baja California Sur, Mexico | HQ596464 | 1,140 | a |
| BT | Lepus californicus | Baja California Sur, Mexico | HQ596465 | 1,140 | a |
| BT | Lepus californicus | Estado de Mexico, Mexico | HQ596463 | 1,140 | a |
| BT | Lepus californicus | Utah, USA | HQ596466 | 1,140 | a |
| BT | Lepus californicus | Lubbock, Texas, USA | U58933 | 1,140 | d |
| BT | Lepus californicus | Bernalillo County, New Mexico, USA | AF010160 | 1,140 | b |
| WT | Lepus townsendii | Utah, USA | HQ596485 | 1,140 | a |
| WT | Lepus townsendii | USA | AY292729 | 1,140 | e |
| WT | Lepus townsendii | Cache Valley, Utah, USA | AF009733 | 702 | e |
| OG | Sylvilagus audubonii | Mexico | HQ596488 | 1,140 | a |
| OG | Romerolagus diazi | Mexico | HQ596487 | 1,140 | a |
We used SEQUENCHER V.5.4.1. (Gene Codes Corp. Ann Arbor, Michigan) to edit and map the nucleotide fragments of the 2 museum samples to a reference sequence. We used the consensus sequence of 5 species of Lepus as the reference: L. alleni, L. californicus, L. callotis, L. flavigularis, and L. townsendii (GenBank accession numbers HQ596458, HQ596463, HQ596468, HQ596475, HQ596485 respectively). We then used the “Muscle algorithm” (Edgar, 2004) as implemented in “Sequencher” to align our reference-based assemblies against the entire data set of sequences downloaded from GenBank. The alignment of these 5 species of Lepus and the museum samples (USNM 93692 and USNM 93694) was used to calculate genetic distance using Kimura’s (1980) two-parameter model. The pairwise distance matrix was generated by the dist.dna function from the ape 3.4 R package (Paradis et al., 2004).
We used MrBayes V.3.2.6 (Ronquist & Huelsenbeck, 2003) and RAxML V.1.5b1 (Silvestro & Michalak, 2012) to determine interspecific relationships based on Bayesian and Maximum-likelihood (ML) inferences, respectively. We selected the GTR+G+I model of sequence evolution, as determined by the corrected Akaike information criterion (AICc) in jModelTest V. 2.1.10 (Darriba et al., 2012). In MrBayes, we performed 4 independent runs, each one consisting of 1 cold and 3 heated chains (1.00, 0.91, 0.83, 0.77, respectively) of 20 million generations sampled every 1,000 generations. We used Tracer V.1.6 (Rambaut et al., 2013) to verify convergence within and between runs and to ensure adequate sampling of all parameters. We removed the first 2,000 trees of the posterior samples as burn-in (mean LnL = -3992; ESS = 16909) and summarized the phylogenetic trees in MrBayes. We generated ML trees in RAxML with the “ML + rapid bootstrap” option, which performs a bootstrap analysis and searches for a best-scoring ML tree. We assessed node support by selecting “automatic bootstopping” (Pattengale et al., 2010) according to the majority rule tree consensus-based criteria (“-N autoMR”). We visualized and edited resulting trees in FigTree V.1.4.2 (Rambaut, 2014) and Adobe Illustrator CC 20.1.0.
Results
Our examination of L. altamirae at the National Museum showed that these animals closely resembled L. callotis callotis specimens from Jalisco. Furthermore, the white flanks, black stripes along the nape, ocherous throats, and dusky rather than black ear tips suggested a closer relationship to L. callotis, L. flavigularis and L. alleni than to L. californicus (Fig. 1).
The concatenated set of primer pairs we designed targeted a 529 base pair fragment of the MT-CYB gene, including 70 polymorphic (informative) sites. Sequence data for museum samples USNM 93692 and USNM 93694 consisted of 5 concatenated fragments of 461 bp (59 polymorphic sites) and 475 bp (57 polymorphic sites), respectively.
Genetic distances within the Lepus genus ranged from 0.017 to 0.095 (Table 3). The distance between samples USNM 93694 and USNM 93692 and L. flavigularis was 0.017 and 0.020 respectively. Whereas the distance of the museum samples and L. californicus was 0.032 USNM 93694 and 0.034 for USNM 93692.
Table 3 Genetic distances between 5 North American Lepus species: Lepus townsendii (LT), Lepus alleni (LA), Lepus californicus (LCA), Lepus callotis (LA), Lepus flavigularis (LF) and L. altamirae: USNM 93692 (92), USNM 93694 (94).
| 92 | 94 | LT | LA | LCA | LC | LF | |
|---|---|---|---|---|---|---|---|
| 92 | - | ||||||
| 94 | 0.007 | - | |||||
| LT | 0.080 | 0.077 | - | ||||
| LA | 0.037 | 0.035 | 0.094 | - | |||
| LCA | 0.035 | 0.032 | 0.080 | 0.022 | - | ||
| LC | 0.040 | 0.042 | 0.095 | 0.045 | 0.037 | - | |
| LF | 0.020 | 0.017 | 0.078 | 0.027 | 0.025 | 0.032 | - |
Bayesian and ML methods produced identical phylogenies with respect to the 5 species of Lepus, the outgroup taxa, and the museum samples. Within the Lepus genus, we recovered 2 distinct monophyletic clades. The first clade (A) corresponded to representatives of L. townsendii. The second clade included 3 separate groups: B, C and D (Fig. 3). L. callotis (group B) appeared as sister group to 4 taxa: L. alleni, and L. californicus (group C), and L. flavigularis and the museum samples (group D). Support for the sister relationship of these 2 groups (C and D) was low in both Bayesian and ML reconstruction methods (posterior probability = 0.67; bootstrap = 48%, respectively).
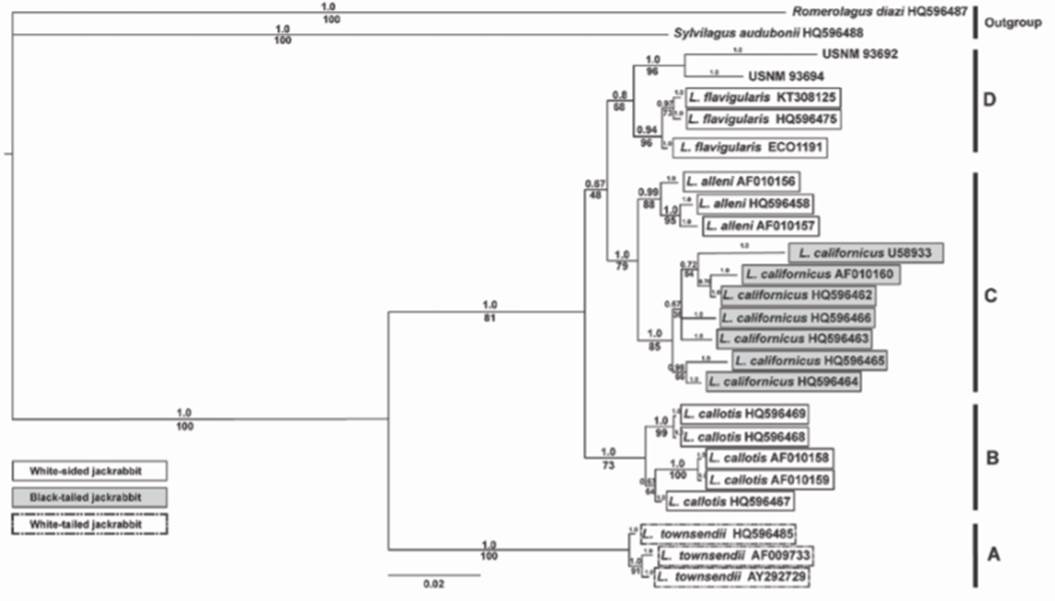
Figure 3 Phylogeny reconstructed by Bayesian and ML algorithms based on MT-CYB sequences using MrBayes and RAxML software programs, respectively, and GTR + G + I as the optimal substitution model. Posterior probability values are above branches and bootstrap support are given below branches. Note the location of USNM 93692 and 93694 as belonging to the white-sided clade with L. flavigularis.
L. alleni and L. californicus clustered together in one clade with high support in both methods (posterior probability = 1.0; bootstrap = 79%), whereas L. flavigularis and the museum samples were separated from these 2 taxa into another group with moderate posterior probability (0.8), and relatively low bootstrap support (68%). Bayesian and ML phylogenetic analyses support a relationship between the museum samples (L. altamirae) and the group of white-sided jackrabbits, L. flavigularis.
Discussion
Our phylogenetic analyses based on MT-CYB sequences found white-sided jackrabbits to be paraphyletic, where L. alleni and L. californicus are more closely related to each other. Our data support the placement of L. flavigularis and the museum samples in a monophyletic group, with L. altamirae more closely related to representatives of a white-sided species of jackrabbit (L. flavigularis) than to black-tailed species (L. californicus). This relationship is supported by our analyses of genetic distance, which show that L. altamirae jackrabbits are closer to L. flavigularis (0.017-0.020) than to any other Lepus species (0.035-0.080).
The placement of L. townsendii as a group separate from the rest of the species of Lepus is consistent with previous studies (Halanych et al., 1999; Lorenzo et al., 2014; Ramírez-Silva et al., 2010). The relationship of L. alleni and L. californicus as sister taxa, with L. callotis as sister to both was also found by Halanych et al. (1999) based on MT-CYB data. However, other phylogenetic analyses based on MT-CYB sequences support the monophyly of white-sided jackrabbits (L. alleni, L. callotis, and L. flavigularis), where L. callotis, and L. flavigularis are more closely related to each other (Lorenzo et al., 2014; Ramírez-Silva et al., 2010).
The relatively low or moderate support for some groups (C and D as sister groups) can be attributed to the limited base pairs of sequence data obtained from ancient museum sample DNA, which is highly degraded and more difficult to analyze than modern high-quality DNA (Leonard, 2008). Resolution and support for phylogenetic reconstruction can be improved by increasing the number of characters (base pairs of DNA analyzed) and genetic markers utilized in the data set (Heath et al., 2008). Although short sequences typically reflect accurate phylogenetic relationships at the species level, low statistical support at many nodes can misrepresent some of the branching patterns (Min & Hickey, 2007).
This study provides an initial estimate of the evolutionary relationship of L. altamirae to other North American Lepus species. A more robust estimation of phylogenetic relationship that reflects the true tree topology with high statistical support is needed for a robust placement of L. altamirae within the white-sided group of jackrabbits.
In order to recover relationships in a more robust manner, a phylogenomic study including additional sampling would be ideal. Whole genome datasets rich in both species and genes reflect the divergence of an organism better than methods based on selected genes and are likely to produce more accurate results (Delsuc et al., 2005; Sims et al., 2009) reconstructing the true tree topology with high support.
Based on the data we obtained in this study, it appears that Nelson’s 1909 hypothesis was correct: L. altamirae is a species of white-sided jackrabbits confined to the tropical and subtropical savannas of southern Tamaulipas. From a conservation perspective, these results are significant because this species may still be extant, as an animal resembling L. altamirae was photographed in Tamaulipas by a trail camera set (remote-triggered camera) in the Área de Protección de Flora y Fauna Laguna Madre y Delta del Río Bravo (572,808 ha) by the Comisión Nacional de Areas Naturales Protegidas (Conanp) and published in social media in 2016 (Fig. 4). Another photograph showing an animal resembling L. altamirae, tagged as black-tailed jackrabbit, was taken on December 14, 2014, in Soto La Marina, Tamaulipas, Mexico and posted on the iNaturalist website (www.inaturalist.org/observations/1139454; Fig. 5).

Figure 4 Putative individual of a white-sided jackrabbit recorded by the Comisión Nacional de Áreas Naturales Protegidas (Conanp) camera on its Área de Protección de Flora y Fauna Laguna Madre y Delta del Río Bravo in Tamaulipas as shown on “Facebook” and “Twitter” in 2016. Note the white flanks and lack of black on ear tips.

Figure 5 Snapshot of a putative individual of a white-sided jackrabbit from Soto La Marina, Tamaulipas, Mexico, posted on the iNaturalist website on December 14, 2014. Note the white flanks, black lines on nape and small area of black on ear tips.
Specimens of L. altamirae have not been collected in more than 100 years and this species may have suffered a reduction in distribution similar to or greater than experienced by L. flavigularis (Lorenzo et al., 2008) and L. callotis, the latter species having been replaced over much of its range by L. californicus (Anderson, 1972; Brown et al., 2018).
From a biogeographic standpoint, it appears more consistent to have a white-sided jackrabbit in tropic-subtropic Tamaulipas. The biogeographic pattern would suggest that white-sided jackrabbits retained a greater distribution in the Americas in the pre-Pleistocene era. Then more recently, climate change and the arrival of the black-tailed jackrabbits have caused isolation of white-sided jackrabbits in tropic-subtropic environments (Lorenzo et al., 2014). Under this hypothesis, there should be a white-sided jackrabbit in both the northeastern and the northwestern American tropics (as well as the Tehuantepec Isthmus). The reinstatement of the L. altamirae species, restores an eastern analog to the western white-sided L. alleni.











 nueva página del texto (beta)
nueva página del texto (beta)


