Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Revista mexicana de biodiversidad
versión On-line ISSN 2007-8706versión impresa ISSN 1870-3453
Rev. Mex. Biodiv. vol.81 no.3 México dic. 2010
Ecología
Genetic structure of a bird–dispersed tropical tree (Dendropanax arboreus) in a fragmented landscape in Mexico
Estructura genética de un árbol tropical dispersado por aves (Dendropanax arboreus) en un paisaje fragmentado en México
Elsa M. Figueroa–Esquivel1, Fernando Puebla–Olivares2, Luis E. Eguiarte3 and Juan Núñez–Farfán1*
1 Laboratorio de Genética Ecológica y Evolución, Departamento de Ecología Evolutiva, Instituto de Ecología, Universidad Nacional Autónoma de México, Apartado postal 70–275, 04510 México, D.F., México. *Correspondent: farfan@servidor.unam.mx
2 Museo de Zoología Alfonso L. Herrera, Facultad de Ciencias, Universidad Nacional Autónoma de México, Apartado postal 70–399, 04510 México, D.F., México.
3 Laboratorio de Evolución Molecular y Experimental, Departamento de Ecología Evolutiva, Instituto de Ecología, Universidad Nacional Autónoma de México, Apartado postal 70–275, 04510 México, D.F., México.
Recibido: 07 octubre 2009
Aceptado: 23 marzo 2010
Abstract
We analyzed the genetic structure of the tropical tree Dendropanax arboreus (Araliaceae) in relation to habitat fragmentation. Genetic variation, structure, and genetic differentiation among populations from Los Tuxtlas tropical rainforest were estimated using ISSRs as molecular markers. DNA from 219 individuals belonging to 9 populations was amplified with 4 primers yielding a total of 75 loci. Adults and juveniles from each population were analyzed to assess the genetic diversity and structure pre and post–fragmentation, respectively. Dendropanax arboreus showed high levels of genetic diversity (h = 0.253) and significant but low genetic differentiation among populations (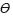 = 0.062). A hierarchical analysis of the gvenetic structure showed that 91.5% of the genetic variation is attributable to individual differences within populations. The average Nei's genetic distance among populations was low (D = 0.034) and genetic distance among pairs of populations increased with geographic distance separating them. Because genetic diversity is similar between adult and juvenile trees at all but 2 populations, we suggest that seed dispersal prevented genetic differentiation and maintains genetic connectivity among fragments and continuous forest populations. Juvenile populations showed a higher genetic differentiation (
= 0.062). A hierarchical analysis of the gvenetic structure showed that 91.5% of the genetic variation is attributable to individual differences within populations. The average Nei's genetic distance among populations was low (D = 0.034) and genetic distance among pairs of populations increased with geographic distance separating them. Because genetic diversity is similar between adult and juvenile trees at all but 2 populations, we suggest that seed dispersal prevented genetic differentiation and maintains genetic connectivity among fragments and continuous forest populations. Juvenile populations showed a higher genetic differentiation (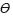 = 0.15) than adult trees, indicating a role of genetic drift via reduced population size.
= 0.15) than adult trees, indicating a role of genetic drift via reduced population size.
Key words: conservation, tropical rain forest, Dendropanax arboreus, genetic structure, habitat fragmentation, ISSR.
Resumen
Se analizó la estructura genética del árbol tropical Dendropanax arboreus (Araliaceae) en relación con la fragmentación del hábitat en la selva tropical de Los Tuxtlas, Veracruz, México. La variación, estructura y diferenciación genética entre poblaciones del bosque continuo y de fragmentos se estimó usando ISSR como marcador molecular. El ADN de 219 individuos de 9 poblaciones se amplificó para 4 primers (75 loci). Se analizaron las poblaciones de árboles adultos y juveniles en cada sitio, representando la diversidad y estructura genética pre y postfragmentación, respectivamente. Dendropanax arboreus mostró altos niveles de variación genética (h = 0.253) y un nivel bajo de diferenciación entre poblaciones 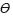 (= 0.062). Un análisis jerárquico de la estructura genética mostró que el 91.5% de la variación genética es atribuible a diferencias individuales dentro de las poblaciones. El promedio de las distancias genéticas de Nei entre las poblaciones fue bajo (D = 0.034), mientras que la distancia genética entre pares de poblaciones se incrementó con la distancia geográfica. En nivel poblacional, el efecto de la fragmentación no es evidente aún, lo cual podría deberse a una alta y efectiva dispersión de semillas por aves. Se sugiere que la dispersión de semillas ha mantenido la conectividad entre las poblaciones de selva continua y fragmentos. Las poblaciones juveniles muestran una diferenciación genética mayor (
(= 0.062). Un análisis jerárquico de la estructura genética mostró que el 91.5% de la variación genética es atribuible a diferencias individuales dentro de las poblaciones. El promedio de las distancias genéticas de Nei entre las poblaciones fue bajo (D = 0.034), mientras que la distancia genética entre pares de poblaciones se incrementó con la distancia geográfica. En nivel poblacional, el efecto de la fragmentación no es evidente aún, lo cual podría deberse a una alta y efectiva dispersión de semillas por aves. Se sugiere que la dispersión de semillas ha mantenido la conectividad entre las poblaciones de selva continua y fragmentos. Las poblaciones juveniles muestran una diferenciación genética mayor (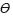 = 0.15) que las poblaciones de adultos, sugiriendo un papel de la deriva génica vía reducción en el tamaño poblacional.
= 0.15) que las poblaciones de adultos, sugiriendo un papel de la deriva génica vía reducción en el tamaño poblacional.
Palabras clave: conservación de una selva tropical, Dendropanax arboreus, estructura genética, fragmentación del hábitat, ISSR.
Introduction
Theory regarding the effects of habitat fragmentation on the genetic structure of populations predicts an increment in the magnitude of genetic divergence among them, and a reduction in gene flow, that in turn may produce genetic drift and increased inbreeding, further isolation, and reduction of population sizes (Hall et al., 1996; Young et al., 1996; Nason et al., 1997; see Lowe et al., 2005). The magnitude and direction of change in ecological and genetic parameters of populations brought about by habitat fragmentation could depend on the magnitude of gene flow among populations (Templeton et al., 1990; Saunders et al., 1991; Chase et al., 1995; Couvet, 2002), and the landscape structure, which influences the movement of genes (Sork et al., 1999).
Pollen dispersal is considered the most important determinant of gene flow reducing the genetic structuring of plant populations (Ellstrand and Elam, 1993). Nevertheless, gene flow via seed dispersal may be more extensive than pollen dispersal in tropical trees (Dick et al., 2008) and more effective in maintaining genetic connectivity among populations inhabiting forest fragments (Bacles et al., 2006), thus preventing differentiation among populations (Nason et al., 1997). Because seed dispersal involves genes from both parents, gene flow through seeds accounts for two–thirds of total genetic neighbourhood size, and is of utmost importance for colonization of new habitats (Hamrick et al., 1993; Hamrick and Nason, 1996; Schnabel et al., 1998; Hamilton, 1999; Gaiotto et al., 2003).
Endozoochorous plant species can exhibit higher gene flow and genetic variation among populations than species with other seed dispersal syndromes (Jordano and Godoy, 2000). Data obtained from bird–dispersed trees and shrubs in both the tropics (e.g., Gibson and Wheelwright, 1995; Loiselle et al., 1995a; Hall et al., 1996; Gauer and Cavalli, 2000; Chung et al., 2000) and temperate forests (Bruederle et al., 1998; Jordano and Godoy, 2000; Bacles et al., 2004), suggest that long–distance seed dispersal is important in preventing genetic drift (Hall et al., 1996). However, the spatial pattern and extent of gene flow by seed dispersal varies greatly among plant species, because dispersers differ greatly in their effectiveness (Loiselle et al., 1995b; Jordano and Godoy, 2000; Figueroa et al., 2009).
This study assessed the genetic structure of the tropical tree Dendropanax arboreus at Los Tuxtlas tropical rain forest, a habitat highly fragmented due to deforestation (Dirzo and García, 1992). Because D. arboreus is dispersed by a great variety of birds in this region, we hypothesized that fragmentation has not exacerbated genetic differentiation among populations (Loveless, 1992), nor has it reduced genetic diversity within populations (Gauer and Cavalli, 2000), since gene flow via seed dispersal by birds is still maintained (Hamrick et al., 1993; Jordano and Godoy, 2000; Bacles et al., 2004).
Materials and methods
Dendropanax arboreus (L.) Decne. et Planch. (Araliaceae) is an evergreen canopy tree (14 – 25 m height). Its geographic distribution ranges from Mexico to South America, occurring at elevations from sea level up to 1500 m asl. In Mexico this tree has a wide distribution in the tropical regions and it is relatively common in the rain forest of Los Tuxtlas. It grows in primary and secondary forests, in soils with good drainage, or near streams (Pennington and Sarukhán, 1998). It produces hermaphrodite flowers (Ibarra–Manríquez, 1985) pollinated by small insects (Bawa et al., 1985). At Los Tuxtlas, the main floral visitors are Trigona bees, Apis mellifera, and Urania moths (E. Figueroa, pers. obs.). An analysis of the genetic structure of D. arboreus in Costa Rica using RAPDs suggests that this tree species is an outcrosser with low genetic differentiation due to long distance gene flow via pollen (Schierenbeck et al., 1997).
Birds are frequent fruit consumers and dispersers of D. arboreus during mid autumn through winter. At Los Tuxtlas, D. arboreus produces large crops of ripe fruits that attract up to 52 bird species, but fruit dispersal depends strongly on members of the Turdidae family and migratory birds. Hylocichla mustelina behaves as a legitimate seed disperser, and accomplishes long–distance seed dispersal in the area (Figueroa et al., 2009). Dendropanax arboreus is a common pioneer species in tropical forests and requires canopy gaps to reach reproductive maturity (Martínez–Ramos, 1985). Because this species produces large fruit crops, it is considered to have good reproduction but poor seedling recruitment (sensu Bongers et al., 1988; Martínez–Ramos and Soto–Castro, 1993).
This research was conducted at the forest of Los Tuxtlas Biological Research Station (LTBS hereafter) and in fragments of rain forest in its vicinity. The LTBS is located within the Los Tuxtlas Biosphere Reserve in southeastern Veracruz, Mexico. The region is of biogeographical importance because it is close to the boreal limit of the high evergreen rain forest in the Americas (Rzedowski, 1963), and it has been heavily deforested over the last 40 years (Dirzo and García, 1992).
Nine populations of D. arboreus were chosen for this study (Fig. 1, Table 1), 3 located in continuous forest (Estación, Laguna, and Estación–Laguna), and 5 in fragments of rain forest of different area (3, 40, 60, and 150 ha). Trees in corridors connecting the fragments (living fences, isolated trees, or riparian remnants; 2.3 ha; , Table 1) were also sampled. Because D. arboreus is a long–lived tree, reproductive individuals in fragments preceded fragmentation and thus the potential impact of fragmentation on genetic variation and structure could be equivocal. For this reason, efforts were made to collect juvenile and seedling individuals in each sampled site to constitute the post–fragmentation generation. Yet, since D. arboreus regenerates preferentially in forest–gaps, young individuals are very rare.
We collected young leaves of all available individuals of D. arboreus in each population (a total of 219 individuals, 151 adults, and 68 juveniles). The minimum number of analyzed individuals was 10 in the population Santa Marta, and the maximum number of individuals was 42 in the Chepe fragment; the average number of individuals per population was 24 (, Table 2).
The leaves were stored, labeled, and frozen in liquid nitrogen and transferred to storage at –70° C. Genomic DNA was extracted using a modified CTAB technique (Doyle and Doyle, 1987). The tissue was ground to a fine powder using liquid nitrogen in a mortar. The quality and concentration of DNA were estimated using 0.7% agarose gels and a λ DNA size marker (Invitrogen 25250–010™). For some samples it was necessary to confirm DNA concentration using a spectrophotometer (Gene Quant Pro; Amersham Biosciencies); all samples were adjusted to a concentration of 10 ng/μl, purified with RNAse (10 mg/ml; SIGMA R4875 ™), and incubated at 37°C for 1 h.
DNA was amplified by PCR using primers of Inter–Simple Sequence Repeats (ISSR; synthesized by Invitrogen): 808 (17 bands, 234–1 186 bp, (AG)8 C); 809 (19 bands, 215–1 360 bp, (AG)8 G); 811 (20 bands, 238–1 520 bp, (GA)8 C); 840 (19 bands, 146–1 236 bp, (GA)8 YT). Amplification reactions were performed in a total volume of 12 μl containing 2 μl of template DNA [20 ng], 4 μl of primer and 6 μl of ReadyMixTM (Taq PCR Reaction SIGMA P4600™). The reactions were performed in a Thermal Cycler (Applied Biosystems 2720), with the following program: 94°C (2 min); 35 cycles (94°C, 30 sec; 46°C, 35 sec; 72°C, 2 min); 72°C (7 min); 4° C. For each primer, controls were included to verify repeatability of results.
Amplified products were electrophoresed in 2% agarose gels with 1X TBE buffer at 120 V for 3.5 h., and stained with ethidium bromide. The pattern of ISSR bands was visualized and photographed under UV light in a transilluminator. Molecular mass of the bands was estimated by comparison with 1 Kb Plus ladder (100 to 12 000 bp; Invitrogen 10787–018™).
Gel scoring was performed manually, with bands were scored as present or absent; a matrix of ISSRs bands was constructed. We assumed that each band represented a single biallelic locus in Hardy–Weinberg equilibrium, and the presence or absence of bands can be transformed into allelic frequencies. For the analysis, we used exclusively data from those individuals whose PCR products were amplified for all primers. The matrix was analyzed with the programs Tools for Population Genetic Analyses (TFPGA v. 1.3, using Lynch and Milligans' correction; Miller, 1997) and Population Genetic Analyses (POPGENE v. 1.31; Yeh et al., 1997). Analyses were performed including all individuals in each population regardless of their life stage, and separate analyses were conducted for adult (pre–fragmentation) and young plants (post–fragmentation).
For each population, we estimated genetic diversity (h), Shannon's index of phenotypic diversity (I), and percentage of polymorphism (% P). To test for significant differences in genetic diversity (h) among populations, we performed ANOVA (one–way), and to compare the mean values of genetic diversity we used the test of LS Fisher (Least Squares Means). Both analyses were performed in the program Data Analysis Software System (STATISTICA v. 6.0; StatSoft, Inc., 2001).
To determine population differentiation of D. arboreus, we used Lynch and Milligan's Fst or the coefficient of coancestry (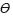 ), because it has been suggested that it is a better estimator for dominant data (Culley, 2009). All values of genetic differentiation have a theoretical minimum of 0 (no genetic divergence) and a theoretical maximum of 1 (fixation of alternative alleles in different populations).
), because it has been suggested that it is a better estimator for dominant data (Culley, 2009). All values of genetic differentiation have a theoretical minimum of 0 (no genetic divergence) and a theoretical maximum of 1 (fixation of alternative alleles in different populations).
We inferred gene flow among populations applying a Bayesian clustering algorithm implemented in the program Structure (Pritchard et al., 2000). After examining various burn–in lengths, we observed that a chain length of 500 000 iterations with 106 Markov chain Monte Carlo (MCMC) was suitable to explore population structure, letting the number of populations (K) to vary between 1 and 10. We estimated the number of clusters according to the procedure of Evanno et al. (2005). We assumed the no–admixture model with allele frequencies correlated among populations (Falush et al., 2003).
The resulting groups from the Structure analysis (Pritchard et al., 2000) were employed to construct a dendrogram by the UPGMA (Unweighted Pair–Group Method with Arithmetic Averaging) method using Euclidean distances. We further used an AMOVA (Program Arlequin v. 2.0; Schneider et al., 2000) to partition genetic variation within and among groups. The level of genetic variation was measured by ΦCT, which refers to differences among groups, among populations within groups by FSC, and within populations (ΦST).
Genetic distances among populations were calculated using Nei's (1972) method. This method directly estimates genetic differences among pairs of populations based on allelic differentiation. The isolation by distance model was tested by correlating the matrices of genetic and geographic distances among populations using a Mantel test implemented in the program TFPGA. The geographical distances were calculated using Google Earth (Image © 2006, TerraMetrics).
Results
Eighty percent of the amplified bands (60 loci, 75 in total) were polymorphic. The percentage of polymorphic bands ranged from 54.7% (Santa Marta site) to 78.7% (Huber site) (, Table 2). The average genetic diversity (h) for all populations was 0.253 ± 0.184, whereas at species level it was 0.280 ± 0.169. The Santa Marta and Huber populations had the lowest and highest values of h and I, respectively, and differed significantly between them. Both populations were in fragmented forest (, Table 2).
At Los Tuxtlas, D. arboreus showed low genetic structuring. Differentiation among populations was significantly different from zero (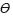 = 0.062; x2 = 696.8; d.f. = 150, P < 0.0001). Genetic differentiation among juveniles of the different populations was 2–fold larger than differentiation among adult populations (juveniles:=
= 0.062; x2 = 696.8; d.f. = 150, P < 0.0001). Genetic differentiation among juveniles of the different populations was 2–fold larger than differentiation among adult populations (juveniles:= 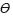 0.15, x2 = 374.5, d.f. = 150, P < 0.0001; adults:=
0.15, x2 = 374.5, d.f. = 150, P < 0.0001; adults:=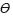 0.08, x2 = 588.1, d.f. = 150, P < 0.0001). Structure analysis detected 2 groups, but these are unrelated to the type of habitat (fragmented or continuous forest). An AMOVA performed on the groups and subgroups derived from Structure analysis detected significant differences between the 2 groups, which accounted for 2.75 % of the genetic variance; subgroups within groups explained 5.7 % of the genetic variance (, Table 3). A dendrogram constructed by UPGMA shows the arrangements of populations (Fig. 2). In general terms, 1 group included individuals from the populations Cerro Borrego, Estación, Playa, and Laguna, whereas the other group included the individuals from Estación–Laguna, Chepe, Huber, and Santa Marta, but each one of these populations included individuals with different percentages of assignment to both groups on the basis of their ancestry (Fig. 3).
0.08, x2 = 588.1, d.f. = 150, P < 0.0001). Structure analysis detected 2 groups, but these are unrelated to the type of habitat (fragmented or continuous forest). An AMOVA performed on the groups and subgroups derived from Structure analysis detected significant differences between the 2 groups, which accounted for 2.75 % of the genetic variance; subgroups within groups explained 5.7 % of the genetic variance (, Table 3). A dendrogram constructed by UPGMA shows the arrangements of populations (Fig. 2). In general terms, 1 group included individuals from the populations Cerro Borrego, Estación, Playa, and Laguna, whereas the other group included the individuals from Estación–Laguna, Chepe, Huber, and Santa Marta, but each one of these populations included individuals with different percentages of assignment to both groups on the basis of their ancestry (Fig. 3).
The average Nei's (1972) genetic distance among all populations was 0.034 ± 0.013 (36 pairwise comparisons). The range of Nei's genetic distances among populations varied from 0.013 to 0.063, and a positive relationship between the geographic and genetic distances was found (Mantel's test: r = 0.7175, P = 0.029). However, this relationship is highly influenced by the Santa Marta population. When this population was removed, the average geographic distance was reduced from 11.97 to 2.87 km (, Table 4), and the relationship between the geographic and genetic distances became not significant (Mantel's test: r = 0.1549, P = 0.262).
Adult vs. Juvenile Individuals
The average genetic diversity (h) for adults was 0.239 ± 0.19, while for juveniles it was 0.199 ± 0.196. The Huber adults and Chepe juveniles had the highest average genetic diversity (Fig. 4). Playa, Estación, and Chepe populations had higher genetic diversity in juvenile than in adult plants (Fig. 4). The Santa Marta and Corredor juveniles had the lowest average genetic diversity. To assess if genetic diversity depended upon sample size of adults (pre–fragmentation) and juvenile plants (post–fragmentation), adult population size was equalled to the juvenile population size of the same location by removing adults at random (Leberg, 2002). Results show that in general, h did not differ between adults and juveniles with the exception of Santa Marta and Corredor, both are populations with lower sample sizes (Fig. 4).
Discussion
Our results show that although the rain forest at Los Tuxtlas region has been drastically reduced and fragmented, Dendropanax arboreus possess high levels of genetic diversity, comparable to other tropical trees dispersed by animals with an outcrossing mating system (Hamrick, 1989; Hamrick and Godt, 1996). However, it is possible that the levels of genetic diversity have not been reduced in Dendropanax populations, because fragmentation is recent compared to the species' life span (Collevatti et al., 2001; Kramer et al., 2008).
Nevertheless, the high levels of genetic variation, the relatively low 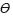 value, the overlap of individuals within the 2 genetic groups, and the low genetic distances among populations, suggest that historically and/or currently, gene flow is high among the studied sites. We sugest that it is possible that contemporary gene flow is still high between fragments and the continuous forest, because D. arboreus has high levels of outcrossing rates and long distance gene flow both via pollen (Schierenbeck et al., 1997) and via seeds (Figueroa et al., 2009). Thus, genetic connectivity among fragments will depend on pollen and seed vectors, the foraging behaviour of dispersers, and the fragmentation matrix (Aguilar et al., 2008; Kramer et al., 2008). No direct estimates of dispersal via pollen are available for this species, but studies with canopy trees showed pollinization over long distances by moths and bees (Bawa, 1990). However, we suggest that the ability of birds as effective seed dispersers throughout the fragmented landscape of Los Tuxtlas is the determining force in maintaining genetic connectivity among populations (Figueroa et al., 2009).
value, the overlap of individuals within the 2 genetic groups, and the low genetic distances among populations, suggest that historically and/or currently, gene flow is high among the studied sites. We sugest that it is possible that contemporary gene flow is still high between fragments and the continuous forest, because D. arboreus has high levels of outcrossing rates and long distance gene flow both via pollen (Schierenbeck et al., 1997) and via seeds (Figueroa et al., 2009). Thus, genetic connectivity among fragments will depend on pollen and seed vectors, the foraging behaviour of dispersers, and the fragmentation matrix (Aguilar et al., 2008; Kramer et al., 2008). No direct estimates of dispersal via pollen are available for this species, but studies with canopy trees showed pollinization over long distances by moths and bees (Bawa, 1990). However, we suggest that the ability of birds as effective seed dispersers throughout the fragmented landscape of Los Tuxtlas is the determining force in maintaining genetic connectivity among populations (Figueroa et al., 2009).
We found comparable levels of genetic diversity in juvenile (post–fragmentation generation) and adults (pre–fragmentation generation), suggesting immigration of alleles via pollen (Eguiarte et al., 1992) or seed dispersal (Bacles et al., 2004). Although a standardization of samples supports this result, other factors, including the effect of the sample size, cannot be ruled out. Nevertheless, the above mentioned is in agreement with the expectation that heterozygosity would not be reduced in long–lived species when fragmentation occurred recently (within less than 50 years) (Aguilar et al., 2008). In the long–term, reductions in genetic diversity by fragmentation would limit the capacity of populations to respond to continuous changes in the environment (Young et al., 1996). In this respect, 2 populations showed a reduction in genetic diversity in juvenile trees, suggesting ongoing genetic drift by habitat fragmentation. In this study, we show that genetic variation in D. arboreus at Los Tuxtlas is mostly contained within populations (91.5%) rather than among populations. This value is similar to those obtained in other studies of outcrossing tree species in fragmented landscapes using dominant markers; for example, 87.9% of genetic diversity in Swietenia macrophylla is maintained within populations (Gillies et al., 1999), and 87.7% within populations of Plathymenia reticulata (Lacerda et al., 2001). These values are consistent with predictions for animal–dispersed species that exhibit a very high proportion of genetic variation within populations (typically >70%; Hamrick et al., 1993).
Dendropanax arboreus at Los Tuxtlas shows low genetic structure, but the magnitude of genetic differentiation among populations is higher than that estimated for populations of this species in Costa Rica (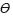 LT= 0.062 vs.
LT= 0.062 vs.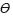 CR = 0.023; Schierenbeck et al., 1997), and for understory palms or other tree species at Los Tuxtlas (e.g., Eguiarte et al, 1992; Pérez–Nasser et al., 1993; Luna et al., 2007; Aguilar, 2008), including pioneer tree species (e.g., Álvarez–Buylla and Garay, 1994). In almost all these plant species, dispersal is by birds, suggesting that the lack of differentiation among fragments at Los Tuxtlas results from the effectiveness of seed dispersal over long distances.
CR = 0.023; Schierenbeck et al., 1997), and for understory palms or other tree species at Los Tuxtlas (e.g., Eguiarte et al, 1992; Pérez–Nasser et al., 1993; Luna et al., 2007; Aguilar, 2008), including pioneer tree species (e.g., Álvarez–Buylla and Garay, 1994). In almost all these plant species, dispersal is by birds, suggesting that the lack of differentiation among fragments at Los Tuxtlas results from the effectiveness of seed dispersal over long distances.
Our results are in agreement with the proposal that fragment boundaries do not represent boundaries of tree populations, because they may benefit from long–distance seed and/or pollen dispersal (Kramer et al., 2008). There is a general tendency for nearby populations to be more genetically related (pattern of isolation by distance; Álvarez–Buylla and Garay, 1994), yet in D. arboreus a clear pattern of isolation by distance was not found.
Results of this study show that the detrimental effects of habitat fragmentation can be retarded for long–lived species and potentially prevented if the lack of a strong population structure in D. arboreus is the result of extensive gene flow by seed dispersers (Figueroa et al., 2009). Common elements of the landscape structure at Los Tuxtlas are the living fences or border strips and riparian remnants that might facilitate current gene flow between fragments and continuous forests. Dendropanax arboreus trees occur at these places which can act as stepping–stones for pollinators and seed dispersers. The insights derived from our study can be of value for regeneration and restoration of fragmented landscapes of highly degraded rain forests like Los Tuxtlas.
The results of population structure for D. arboreus are complex. Perhaps they indicate that the populations in fragments were derived from a much larger population that has mostly disappeared. An alternative possibility is that the populations were derived from another unstudied population in the recent past. Another explanation is that as a pioneer species, D. arboreus grows only in open sites such as gaps created by tree falls inside natural or pristine forest, where the populations are scattered groups of even aged individuals, generating a complex metapopulation structure. To solve these problems, direct estimates of gene flow via pollen and seeds, and a detailed phylogeographic analysis using maternal (chloroplast) and codominant analyses throughout its distribution would be critical. We are advancing to obtain this information.
Acknowledgements
We thank to 2 anonymous reviewers for valuable comments on this manuscript. Financial support was provided by a CONACyT (SEMARNAT–CONACYT– 0355) and PAPIIT, UNAM (IN–224908) to J. Núñez–Farfán. We are grateful to Laura Márquez–Valdelamar and Fabiola Ramírez for technical advice, and to G. García and Erika Aguirre–Planter for comments on the manuscript. We thank Braulio Gómez Chagala, S. Cuartas–Hernández and A. Montero for valuable field assistance, and Rosamond Coates for advice regarding Dendropanax. E. Figueroa thanks CONACYT (Scholarship 124859) and the Posgrado en Ciencias Biológicas, UNAM.
Literature cited
Aguilar, A. B. 2008. Efecto de la fragmentación del hábitat en la genética poblacional de la palma Chamaedorea alternans (Wendl.) Arecaceae en la selva tropical de Los Tuxtlas, Veracruz, México. Tesis, Facultad de Ciencias, Universidad Nacional Autónoma de México. México, D.F. 61 p. [ Links ]
Aguilar, R., M. Quesada, L. Ashworth, Y.Herrerias–Diego and J. Lobo. 2008. Genetic consequences of habitat fragmentation in plant populations: susceptible signals in plant traits and methodological approaches. Molecular Ecology 17:5177–5188. [ Links ]
Álvarez–Buylla, E. R. and A. A. Garay. 1994. Population genetic structure of Cecropia obtusifolia, a tropical pioneer tree species. Evolution 48:437–453. [ Links ]
Bacles, C., A. Lowe and R. Ennos 2004. Genetic effects of chronic habitat fragmentation on tree species: the case of Sorbus aucuparia in a deforested Scottish landscape. Molecular Ecology 13:573–584. [ Links ]
Bacles, C., A. Lowe and R.Ennos. 2006. Effective seed dispersal across a fragmented landscape. Science 311:628. [ Links ]
Bawa, K. S. 1990. Plant–pollinator interactions in tropical rain forest. Annual Review of Ecology and Systematics 21:399–422. [ Links ]
Bawa, K. S., D. R. Perry. and H. Beach. 1985. Reproductive biology of tropical lowland rain forest trees. II. Pollination Systems. American Journal of Botany 72:346–356. [ Links ]
Bongers, F., J. Popma, J. Meave del Castillo and J. Carabias 1988. Structure and floristic composition of the lowland rain forest of Los Tuxtlas, Mexico. Vegetatio 74:55–80. [ Links ]
Bruederle, L., D.Tomback, K. Kelly and R. Hardwick 1998. Population genetic structure in a bird–dispersed pine Pinus albicaulis (Pinaceae). Canadian Journal of Botany 76:83–90. [ Links ]
Chase, M., D. Boshier and K. Bawa 1995. Population genetics of Cordia alliodora (Boraginaceae), a neotropical tree. I. Genetic variation in natural populations. American Journal of Botany 82:468–475. [ Links ]
Chung, M., M. Y. Chung, G. Oh. and B. Epperson. 2000. Spatial genetic structure in a Neolitsea sericea population (Lauraceae). Heredity 85:490–497. [ Links ]
Collevatti, R., D. Grattapaglia and J. Hay 2001. Population genetic structure of the endangered tropical tree species Caryocar brasiliense, based on variability at microsatellite loci. Molecular Ecology 10:349–356. [ Links ]
Couvet, D. 2002. Deleterious effects of restricted gene flow in fragmented populations. Conservation Biology 16:369–376. [ Links ]
Culley, T. 2009. Population Genetic Analysis of ISSR Data. http://bioweb.ad.uc.edu/faculty/culley/Protocols/Population%20Genetic%Analysis%20of%20ISSR&20Data Access: 01.VIII.2009. [ Links ]
Dick, C., O Hardy, A. Jones and R. Petit. 2008. Spatial scales of pollen and seed–mediated gene flow in tropical rain forest trees. Tropical Plant Biology 1:20–33. [ Links ]
Dirzo, R. and M.García. 1992. Rates of deforestation in Los Tuxtlas, a neotropical area in southeast Mexico. Conservation Biology 6:84–90. [ Links ]
Doyle J. J. and J. L. Doyle. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19:11–15. [ Links ]
Eguiarte, L. E., N. Pérez–Nasser and D. Piñero. 1992. Genetic structure, outcrossing rate and heterosis in Astrocaryum mexicanum (tropical palm): implications for evolution and Conservation. Heredity 69:217–228. [ Links ]
Ellstrand, N. and D. Elam. 1993. Population genetic consequences of small population size: implications for plant conservation. Annual Review of Ecology and Systematics 24:217–242. [ Links ]
Evanno, G., S. Regnaut, and J. Goudet. 2005. Detecting the number of clusters of individuals using the software Structure: a simulation study. Molecular Ecology 14:2611–2620. [ Links ]
Falush, D., M, Stephens and J. K.Pritchard, 2007. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molecular Ecology Notes 7:574–578. [ Links ]
Figueroa–Esquivel, E. M., F. Puebla–Olivares, H. Godínez–Álvarez, and J. Núñez–Farfán. 2009. Seed dispersal effectiveness by understory birds on Dendropanax arboreus in a fragmented landscape. Biodiversity Conservation. DOI 10.1007/s10531–009–9645–z. [ Links ]
Gaiotto, F., D. Grattapaglia and R. Vencovsky. 2003. Genetic structure, mating system, and long–distance gene flow in heart of palm (Euterpe edulis Mart.). Heredity 94:399–406. [ Links ]
Gauer L. and S. Cavalli. 2000. Genetic variation in natural populations of maté (Ilex paraguariensis A. St.–Hi., Aquifoliaceae) using RAPD markers. Heredity 84:647–656. [ Links ]
Gibson, P. and N. Wheelwright. 1995. Genetic structure in a population of a tropical tree Ocotea tenera (Lauraceae): influence of avian seed dispersal. Oecologia 103:49–54. [ Links ]
Gillies, A. C. M., J. P. Cornelius, A. C. Newton, A. Navarro, M. Hernández, and J.Wilson. 1999. Genetic variation in Costa Rica populations of he tropical timber species Cedrela odorata L., assessed using RAPDs. Molecular Ecology 6:1133–1145. [ Links ]
Hall, P., S. Walker and K. Bawa.1996. Effect of forest fragmentation on genetic diversity and mating system in a tropical tree Pithecellobium elegans. Conservation Biology 10:757–768. [ Links ]
Hamilton, M. 1999. Tropical tree gene flow and seed dispersal. Nature 401:129–130. [ Links ]
Hamrick, J. 1989. Isozymes and the analysis of genetic structure in plant populations. In Isozymes in plant biology, D. Soltis and Soltis P.(eds.). Dioscorides, Oregon. p. 87–105. [ Links ]
Hamrick, J., D. Murawski and J. Nason, 1993. The influence of seeds dispersal mechanisms on the genetic structure of tropical tree populations. Vegetatio 107/108:281–297. [ Links ]
Hamrick, J. and M. Godt. 1996. Effects of life history traits on genetic diversity in plant species. Philosophical Transactions: Biological Sciences 351:1291–1298. [ Links ]
Hamrick, J. and J. Nason. 1996. Consequences of dispersal in plants. In Population dynamics in ecological space and time, O. Rhodes, R. Chesser and M. Smith (eds.). Chicago Press. p. 203–236. [ Links ]
Ibarra–Manríquez, G. 1985. Estudios preliminares sobre la flora leñosa de La Estación de Biología Tropical Los Tuxtlas .Tesis, Facultad de Ciencias, Universidad Nacional Autónoma de México. México, D.F. 265 p. [ Links ]
Image. 2006. Google Earth TerraMetrics version 4.0 http://earth.google.com 01.I.2009 [ Links ]
Jordano, P. and J. Godoy. 2000. RAPD variation and population genetic structure in Prunus mahaleb (Rosaceae), an animal–dispersed tree. Molecular Ecology 9:293–1305. [ Links ]
Kramer, A. T., J. L.Ison, M. V.Ashley and H. F. Howe. 2008. The paradox of forest fragmentation genetics. Conservation Biology 22:878–885. [ Links ]
Lacerda, D. R., D. P Acedo M.., J. P. Lemos Filho and M. B. Lovato. 2001. Genetic diversity and structure of natural populations of Plathymenia reticulata (Mimosoideae), a tropical tree from the Brazilean Cerrado. Molecular Ecology 10: 1143–1152. [ Links ]
Leberg, P. L. 2002. Estimating allelic richness: effects of sample size and bottlenecks. Molecular Ecology 11:2445–2449. [ Links ]
Loiselle, B. A., V. L. Sork, J. Nason and C. Graham. 1995a. Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). American Journal of Botany 82:1420–1425. [ Links ]
Loiselle, B. A., V. L. Sork, J. Nason and C. Graham. 1995b. Comparison of genetic variation in bird–dispersed shrubs of a tropical wet forest. Biotropica 27:487–494. [ Links ]
Loveless, M. D. 1992. Isozyme variation in tropical trees: patterns of genetic organization. New Forest 6:67–94. [ Links ]
Lowe, A., D. Dossier, M. Ward, C. Bacles and C. Navarro. 2005. Genetic resource impacts of habitat loss and degradation; reconciling empirical evidence and predicted theory for neotropical trees. Heredity 95:255–273. [ Links ]
Luna, R., B. K.Epperson and K.Oyama. 2007. High levels of genetic variability and inbreeding in two Neotropical decidious palms with contrasting life histories. Heredity 99:466–476. [ Links ]
Martínez–Ramos, M. 1985. Claros, ciclos vitales de los árboles tropicales y regeneración natural de las selvas altas perennifolias. In Investigaciones sobre la regeneración de selvas altas en Veracruz, México, vol. II., A. Gómez–Pompa and S. del Amo R. (eds.). Alhambra Mexicana, México, D.F. p 191–239. [ Links ]
Martínez–Ramos, M. and A. Soto–Castro. 1993. Seed rain and advanced regeneration in a tropical rain forest. Vegetatio 107/108:299–318. [ Links ]
Miller, M. P. 1997. Tools for population genetic analyses (TFPGA 1.3): a windows program for the an allozyme and molecular population genetic data, http://www.bioweb.usu.edu/mpmbio/index.htm [ Links ]
Nason, J., P. Aldrich and J. Hamrick. 1997. Dispersal and the dynamics of genetic structure in fragmented tropical tree populations. In Tropical forest remnants, W. F. Laurance and R. O. Bierregaard (eds.). The University of Chicago Press, Illinois. p 304–320. [ Links ]
Nei, M. 1972. Genetic distance between populations. American Naturalist 106:283–292. [ Links ]
Pennington, T. D. and J. Sarukhán. 1998. Árboles tropicales de México. Manual para la identificación de las principales especies, segunda edición, Universidad Nacional Autónoma de México and Fondo de Cultura Económica, México, D.F. 521 p. [ Links ]
Pérez–Nasser, N., L. E. Eguiarte and D. Piñero. 1993. Mating system and genetic structure of the distylous tropical tree Psychotria faxlucens (Rubiaceae). American Journal of Botany 80:45–52. [ Links ]
Pritchard, J., M. Stephens and P. Donnelly. 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [ Links ]
Rzedowski, J. 1963. El extremo boreal del bosque tropical siempreverde en Norteamérica continental. Vegetatio 11:173–198. [ Links ]
Saunders, D., R. Hobbs and C. Margules. 1991. Biological consequences of ecosystem fragmentation: a review. Conservation Biology 5:18–32. [ Links ]
Schierenbeck, K., M. Skupski, D.Lieberman and M. Lieberman. 1997. Population structure and genetic diversity in four tropical tree species in Costa Rica. Molecular Ecology 6:137–144. [ Links ]
Schnabel, A., J. Nason and J. Hamrick.1998. Understanding the population genetic structure of Gleditsia triacanthos L.: seed dispersal and variation in female reproductive success. Molecular Ecology 7:819–832. [ Links ]
Schneider, S., D. Roessli and L. Excoffier. 2000. Arlequin: an integrated software package for Population Genetics Data Analysis. Version 2.0. Genetics and Biometry Laboratory, Department of Anthropology, University of Geneva. [ Links ]
Sork, V., J., Nason, D. Campbell and J. Fernandez. 1999. Landscape approaches to historical and contemporary gene flow in plants. Trends in Ecology and Evolution 14:219–224. [ Links ]
StatSofft, Inc. 2001. Data analysis software system version 6.0 (Statistica). [ Links ]
Templeton, A., K. Shaw, E. Routman and S. Davis. 1990. The genetic consequences of habitat fragmentation. Annals of the Missouri Botanical Garden 77:13–27. [ Links ]
Yeh, F., R.Yang , T. Boyle, Z.Ye and J. Mao. 1997. POPGENE, the user friendly shareware for population genetic analysis. Molecular Biology and Biotechnology Center, University of Alberta, Edmonton. [ Links ]
Young, A., T. Boyle and T. Brown. 1996. The population genetic consequences of habitat fragmentation for plants. Trends in Ecology and Evolution 11:413–418.v [ Links ]














