Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Similares em
SciELO
Similares em
SciELO
Compartilhar
Revista mexicana de biodiversidad
versão On-line ISSN 2007-8706versão impressa ISSN 1870-3453
Rev. Mex. Biodiv. vol.80 no.1 México Abr. 2009
Ecología
Contrasting genetic structure in two codistributed freshwater fish species of highly seasonal systems
Estructura genética contrastante en dos especies codistribuidas de peces de agua dulce que habitan sistemas marcadamente estacionales
Ella Vázquez–Domínguez1,*, Angélica Hernández–Valdés1, Aliet Rojas–Santoyo1 and Luis Zambrano2
1 Departamento de Ecología de la Biodiversidad, Instituto de Ecología, Universidad Nacional Autónoma de México. Apartado postal 70–275, Ciudad Universitaria, 04510, México D.F., México.
2 Departamento de Zoología, Instituto de Biología, Universidad Nacional Autónoma de México. Apartado postal 70–153, Ciudad Universitaria, 04510, México D.F., México.
*Correspondencia:
evazquez@ecologia.unam.mx
Recibido: 02 abril 2008
Aceptado: 28 julio 2008
Abstract
Given the seasonal nature of ecosystems such as permanent sinkholes ('cenotes') and temporary wetlands, their fish fauna experience yearly local extinction and colonization processes, with strong fluctuations in population size. We evaluated the genetic diversity, population genetic structure and degree of isolation of populations of Poecilia orri and Gambusia yucatana among and within wetlands and cenotes. We also assessed some abiotic characteristics of the water bodies and their potential relationship with average genetic diversity. Both species showed low genetic diversity, but this was twice as low in P. orri. Populations of G. yucatana showed no genetic structure, whereas those of P. orri did. The genetic divergence results were consistent with isolation between cenotes and wetlands, where the different types of water bodies had a distinctive genetic composition. We suggest that our genetic diversity and differentiation results are associated with the successive, seasonal–yearly population size shrinkage and expansion events (i.e. extinction and recolonization) that occur in these systems, and also with the environmental tolerance, body size, and reproduction characteristics of both species. Our results show how these codistributed species can have markedly different genetic structuring and diversity, most likely determined by their particular biological and ecological characteristics, and provide baseline information for future studies.
Key words: cenotes, Gambusia yucatana, Mexico, microsatellites, mitochondrial DNA, Poecilia orri, wetlands.
Resumen
Dada la naturaleza estacional de ecosistemas como cenotes permanentes y humedales temporales, su fauna íctiológica experimenta procesos locales anuales de extinción y colonización, con grandes fluctuaciones en el tamaño poblacional. Evaluamos la diversidad y estructura genética y el grado de aislamiento de las poblaciones de Poecilia orri y Gambusia yucatana dentro y entre humedales y cenotes. También evaluamos algunas características abióticas de los cuerpos de agua y su relación potencial con la diversidad genética. Ambas especies mostraron baja diversidad genética, pero ésta fue 2 veces más baja en P. orri. Las poblaciones de G. yucatana no mostraron estructura genética, mientras que las de P. orri estuvieron estructuradas y diferenciadas. Los resultados de divergencia genética fueron consistentes con aislamiento entre cenotes y humedales, donde los diferentes tipos de cuerpo de agua tuvieron una composición genética distintiva. Sugerimos que la diversidad y diferenciación genética observadas están asociadas con los eventos sucesivos, estacionales, de disminución y expansión del tamaño poblacional (i.e. extinción y recolonización) que ocurren en estos sistemas; asimismo, asociadas con la tolerancia ambiental, el tamaño corporal y las características de reproducción de las 2 especies. Nuestros resultados muestran cómo estas 2 especies codistribuidas pueden tener una estructura y diversidad genética marcadamente diferentes, probablemente determinada por sus características biológicas y ecológicas particulares, y proveen a su vez información fundamental para estudios futuros.
Palabras clave: cenotes, Gambusia yucatana, México, microsatélites, ADN mitocondrial, Poecilia orri, humedales.
Introduction
Coastal wetlands are characterized by highly variable and unpredictable hydrological regimes, in tight association with rainfall seasonality. In tropical and subtropical freshwater coastal ecosystems, rainfall–driven hydrology is a key factor that regulates water level and flooding (Trexler et al., 2005; Rodríguez and Pizarro, 2007). Particularly in wetlands of the Caribbean region, seasonal hydrological conditions yield marked fluctuations in habitat area between dry and wet seasons. Such seasonal dynamics result in a spatially and temporally fragmented ecosystem, in which permanent water bodies (e.g. lagoons) and karstic aquatic habitats called sinkholes and locally known as cenotes, are found along with intermittently flooded and partially or completely dry ones (e.g. wetlands, temporary cenotes, solution holes) (Snyder et al., 1990; Zambrano et al., 2006). This seasonal drying causes mortality of fishes that fail to locate aquatic refuges, leading to disturbance–dominated population dynamics (Trexler et al., 2005; Zambrano et al., 2006).
The Yucatán Peninsula in south–eastern Mexico is a low and relatively flat plain of porous limestone, where rainwater rapidly infiltrates into the ground, yielding few surface–water drainages and extensive subsurface drainage. This subterranean drainage network is characterized by cenotes, as well as by extensive areas of seasonally flooded, fresh– and brackish–water wetlands (Hall, 1936; Schmitter–Soto et al., 2002; see Zambrano et al., 2006 for a description). Despite the wide distribution and great extent of wetlands in southern Mexico, these systems have received little study. In particular, information about fish species in this region is rather limited (Schmitter–Soto, 1999, and references therein), little is known about the structure of fish populations and communities (Zambrano et al., 2006), and to our knowledge there is only 1 genetic study in the region (for Gambusia yucatana; Barona and Espinasa, 2004).
The largest protected area of these aquatic ecosystems in Mexico is the Sian Ka'an Biosphere Reserve, in the State of Quintana Roo. One of the most abundant fish families in the reserve is Poeciliidae (Schmitter–Soto and Gamboa Pérez, 1996; Zambrano et al., 2006). Within this family, 2 species, Poecilia orri, Fowler 1943 and Gambusia yucatana, Regan 1914, are present in most aquatic systems (Zambrano et al., 2006). Seasonality imposes a dichotomy of aquatic systems at Sian Ka'an: the shallow, temporary habitats are dynamic in water level and dry in most years, while deeper systems are relatively stable throughout the year. An important feature of these ecosystems is that during the dry season, isolated sites that maintain some water level serve as refugia for fish species, while during most wet seasons, flooding unites many of them into a continuous pool of aquatic habitat, facilitating dispersal and colonization of fishes into formerly dry habitats (Zambrano et al., 2006). Consequently, the fish fauna experiences yearly local extinction and colonization processes, which structures populations at different temporal and spatial scales.
The genetic structure and extent of population subdivision in patchy or fragmented habitats is determined, among others, by their degree of isolation (Young and Clarke, 2000), by characteristics such as dispersal, generation time or mating system, and by the amount and direction of gene flow (Frankham et al., 2002). Biological and ecological features of species, and geographic distance, topography and environmental conditions, are also important factors structuring populations (Vázquez–Domínguez et al., 1999, 2002; Matoq et al., 2000; Loew et al., 2005). The wetland system in Sian Ka'an is highly intermittent, in which populations are subject to fragmentation during the dry season, with a consequent high mortality. As a general rule, average levels of genetic diversity are expected to be lower in fragmented populations compared with continuously distributed ones, mainly because genetic drift has a higher effect when local effective population size is reduced (Wright, 1931; Slatkin, 1987).
In general, there are few studies evaluating the population genetics and differentiation of freshwater species in seasonal or unstable ecosystems (Mcelroy et al., 2003; Ostergaard et al., 2003; Mamuris et al., 2005). We therefore designed this study to provide baseline information on the genetic diversity, genetic population structure and degree of isolation of P. orri and G. yucatana populations, among and within temporary wetlands and permanent cenotes. We also assessed some abiotic characteristics of the water bodies and their potential relationship with average genetic structuring. Finally, we suggest how some biological and ecological features of these species might be associated with the observed genetic diversity and population differentiation.
Material and methods
Study site and sampling. The Sian Ka'an Biosphere Reserve is located in the State of Quintana Roo, Mexico (20°07'48''N – 18°54'00''N and 88°12'12''W – 87°24'36'' W). Individuals of P. orri and G. yucatana were trapped as part of a continuing work evaluating the community ecology, trophic relationships, genetic diversity and conservation of the fish fauna in the reserve (Zambrano et al., 2006). The study area in the central region of the reserve is based on a hydrological gradient running west to east from inland towards the coast, and along a secondary road approximately 40 km long (Fig. 1a). Sampling was done during wet (September 2001 and November 2002) and dry seasons (March 2002 and April 2003), during an average of 10 days each visit, in 5 temporary (seasonally flooded) and 4 permanent (non–seasonal) water bodies within the reserve (Fig. 1a). Permanent cenotes (LEN, LES, LIM, CeST; see Table 1, Fig. 1a) were sampled in both seasons, while the temporary wetlands PET, LIR, PREM and MAR were sampled only during the wet season; a temporary cenote (SLVA) was sampled only once because it failed to fill up during subsequent wet seasons. As a result of this seasonality and because the study species were abundant in some aquatic habitats, but rather scarce in others (especially cenotes), we have unequal and some very low sample sizes (LES and LIM). We analyzed 56 P. orri and 68 G. yucatana from 7 water bodies per species (Table 1).
Hydrological conditions were characterized in each site to describe the physicochemical features of each water body; basic limnetic data on 6 variables (depth, temperature, pH, dissolved oxygen, turbidity and salinity), were obtained during the morning (10:00–12:00 hrs) with a multiparametric Quanta–Hydrolab unit. Measurements were made at the surface and at 30 cm depth.
Microsatellite typing. Whole genomic DNA was isolated from muscle tissue using the AquaPure Genomic DNA kit (Biorad Laboratories) following the manufacturer's protocol and quantified by spectrophotometry. We surveyed 7 potential microsatellite loci primers developed in the species Gambusia affinis (Spencer et al., 1999): Gafu2, Gafu3, Gafu4, Gafu6 and Gafu7, and G. holbrooki (Zane et al., 1999): Mf–6 and Mf–13; unfortunately the latter were monomorphic for both species studied, while 3 of the former were polymorphic and amplified reliably in P. orri (Gafu3, Gafu4, Gafu6) and G. yucatana (Gafu3, Gafu4, Gafu7).
We amplified the microsatellite loci using polymerase chain reaction (PCR) in a 15 µl reaction volume containing the following: approximately 60 ng template DNA, 0.2 mM each dNTP, 10 x reaction PCR buffer (200 mM Tris–HCl pH 8.4, 500 mM KCl) and 0.45 U Taq Platinum polymerase (Invitrogen); concentration of each primer (0.6 and 0.8 µM) and of MgCl2 (2.5, 3.5 and 3.7 mM) varied depending on primer and species. Amplifications were carried out in a PTC–100 thermal cycler (M.J. Research) as follows: 90 s at 94°C, then 30 cycles of 40 s at 94°C, 40 s at annealing temperature (66°C or 67°C), and a last 60 s at 72°C. We always used negative and positive controls to avoid unspecific amplification and to ensure that amplification profiles were consistent and reproducible. Amplification was done by electrophoresis in 6% acrylamide vertical gels at 500 V for approximately 3 hours. We visualized microsatellite bands by a standard silver staining technique (Sambroock et al., 1989), comparing microsatellite size to molecular weight standards (10 bp ladder; Invitrogen), and performed repetitions on different gels of every sample to assure reproducibility. We digitized the stained acrylamide gels and performed the analysis and genotyping of each sample using LabWorks version 4.5 (Ultra Violet Products).
DNA sequencing. Given that only P. orri showed significant genetic structure (see Results), we selected a subgroup of samples of this species from 6 water bodies that encompassed 3 permanent cenotes: LES (2 individuals), LIM (3) and CeST (7) and 3 temporary wetlands: LIR (3), MAR (5) and PET (4). These samples were sequenced using a fragment of the mitochondrial DNA D–loop region, which was amplified with the primers L15926 (TCAAAGCTTACACCAGTCTTGTAAACC; Kocher et al., 1989) and H16498 (CCTGAAGTAGGAACCAGAT; Meyer et al., 1990). Amplification was done in a 50 µl reaction volume containing the following: approximately 100 ng template DNA, 0.1 mM each dNTP, 10 x reaction PCR buffer (200 mM Tris–HCl pH 8.4, 500 mM KCl), 0.3 U Taq Platinum polymerase, concentration of each primer was 0.4 and 0.6 µM and of MgCl2 was 3.0 and 3.3 mM, the value varying depending on the quality of the DNA sample. We carried out amplifications in a PTC–100 thermal cycler as follows: 180 s at 94°C, then 35 cycles of 70 s at 94°C, 90 s at annealing temperature (53 or 54°C) and a final 150 s at 72°C. We sequenced the purified samples with a 3730xl DNA analyzer.
Statistical analyses. All the following analyses were performed for P. orri and G. yucatana separately, where populations correspond to the different sampling sites for each species.
Genetic diversity. We calculated genetic diversity as the observed (na) and effective (ne) number of alleles, observed (H0) and expected heterozygosity (HE; Levene 1949) and Nei's genetic diversity (HNei; Nei, 1973) for each locus and across loci per site, using the program PopGene v1.31 (Yeh et al., 1997). The p–values were determined by applying the sequential Bonferroni correction for multiple tests (Rice, 1989) over each locus within each population (p < 0.05).
To discern possible associations between genetic diversity and habitat variation (Brouat et al., 2004), we performed canonical correspondence analyses (CCA) using the program Brodgar 2.5.1 (Highland Statistics). CCA incorporates both ordination and multiple regression techniques for direct analysis of the relationship between multivariate data, and assumes a unimodal relationship between dependent and independent variables. For this, variation (among populations) of the intrapopulation diversity was inferred from the variation of the genetic diversity by loci (HNei) as dependent variables, whereas independent variables expected to influence genetic diversity were represented by the 6 limnetic variables measured per water body in both species (Table 1). Values were ln–transformed to obtain normality when necessary.
Population genetic structure and differentiation. We assessed the level of genetic differentiation and population structure by estimating Wright's FST (Slatkin, 1995) and Nei's minimum genetic distance (Dm; Nei 1973) between each sampling site using Arlequin v3.00 (Excoffier et al., 2005) and PopGene, respectively. We did an estimation of the number of migrants per generation (Nm) between sampling sites using M = 1– FST/2FST, where M = 2Nm for diploid populations. We also tested for isolation by distance with a Mantel test based on the geographic distances (in km) and the genetic distances, with 10,000 permutations, with both tests being carried out with Arlequin. Finally, to detect the degree of similarity of the sampling sites based on the species' genotypes, we did a factorial correspondence analysis of the microsatellite data, which graphically projects the individuals in the factor space defined by the similarity of their allelic states, with Genetix 4.05 (http://www.genetix.univ–montp2.fr/genetix/genetix.htm).
Diversity and structure of P. orri based on mitochondrial DNA. We performed sequence alignment with ClustalX v1.53b (Thompson et al., 1997) and edited manually with the Bioedit sequence alignment editor. We grouped bases by homologous codons to obtain an unambiguous alignment, using a published P. orri D–loop fragment (Ptacek and Breden, 1998). We performed Fu and Li's (1993) D– and F–tests of neutrality using the DnaSP 4.10.3 software (Rozas et al., 2003). We did a Maximum Parsimony (MP) analysis with a heuristic search with 100 random additions and implementing the tree bisection–reconnection (TBR) branch–swapping, with PAUP v4.0b10 (Swofford, 2003) and using Poecilia butleri as an outgroup (GenBank number AF080504). Bootstrap values were based on 1,000 replicates and a majority–rule consensus tree was estimated.
We measured genetic diversity by estimating the number of segregating sites (S) and values of haplotype (h) and nucleotide diversity (π) using DnaSP. We evaluated levels of differentiation among populations by estimating the average number of nucleotide substitutions per site (nucleotide divergence, Dxy; Nei, 1987) between water bodies. Because of its small variance and because it does not assume that populations have reached equilibrium, this is a useful index for populations that are spatially close. Also, we constructed a minimum haplotype network under statistical parsimony using the program TCS 1.20 (Clement et al., 2000). Finally, we performed an analysis of molecular variance (AMOVA; Excoffier et al., 1992) to measure population subdivision in P. orri and to estimate the distribution of genetic variation within versus among the 6 water bodies (ΦST, significance level α = 0.05, 10,000 permutations), as implemented in Arlequin.
Results
A high number of alleles per locus was observed, with a total of 27 for P. orri and 30 for G. yucatana. The number of observed and effective alleles across loci per sampling site ranged from na = 3–15 and ne = 1.0–4.1, respectively for P. orri, and na = 6–18 and ne = 1.8–3.9 for G. yucatana (Table 2). P. orri had the highest number of alleles in sites LIR (14) and MAR (15), while G. yucatana had a high number of alleles at all sites, with the lowest number in LES (6). A high number of private alleles (exclusive of a species) was observed: 41% in P. orri, where sites MAR (4) and LIR (3) had the highest number, and 27% in G. yucatana (highest in PET with 3). The observed and expected heterozygosity (without considering LES because it had only 1 allele; see Table 2) across loci for each population ranged between 0–0.222 and 0.178–0.814, respectively for P. orri (mean HO = 0.113, HE = 0.718), while mean HNei was 0.711 (range = 0.148–0.752). G. yucatana showed higher values for H0 = 0.250 (0.159–0.600), and similar for HE = 0.706 (0.422–0.800) and HNei = 0.701 (0.352–0.720).
The first axis in the canonical correspondence analysis explained 96% of the variation in genetic diversity in P. orri, and the eigenvalues indicated that the first axis was considerably more important than the second (0.255 versus 0.012). Distinctively, the variable depth was along this first axis and significantly explained 17% of the total variation (F = 8.096; p = 0.001), while the other 5 limnetic variables were not significant (p > 0.05). G. yucatana showed a less clear pattern for the CCA: the first axis explained 76% of the genetic variation, with eigenvalues of 0.078 for the first axis and 0.025 for the second axis. Three variables were significant (p < 0.05), salinity, oxygen and turbidity, although they explained a rather small percentage (< 3%) of the total variation.
Significant genetic differentiation between some sampling sites was observed with FST for P. orri (Table 3): 17 out of 21 possible pairwise comparisons were significantly different in P. orri; differentiation values ranged from 0.039 to 0.837 and number of migrants from 0.10 to 1.92. A unique high number of migrants (12.3) was observed between the 2 closest temporary wetlands (MAR–PREM, 0.1 km apart, see Table 3), which also had the lowest differentiation value (0.039). The lowest number of migrants and highest differentiation value (0.837) was between 2 permanent cenotes (LES–LIM; 13.1 km; Table 3, Fig. 1). In addition, genetic distance values ranged from 0.048 to 0.631 in P. orri and 0.078 to 0.253 in G. yucatana. The greatest genetic distance values in P. orri were observed between 2 pairs of permanent cenotes (LES–CeST and LES–LIM), and also between LES and the temporary wetland LIR. The lowest values were observed between the 2 closest temporary wetlands (MAR–PREM) (Table 3, Fig. 1). This pattern is in general consistent, in which closer sites had more migrants, were less differentiated and had smaller genetic distances, and vice versa. In contrast, G. yucatana had only 5 significant FST comparisons, values (0.058–0.262) were markedly lower than those observed for P. orri, and the number of migrants were comparatively higher (1.31–8.78). Permanent cenotes showed both high and low genetic distance values when compared to distant temporary water bodies, and low values were also observed between close as well as distant sites (Table 3).
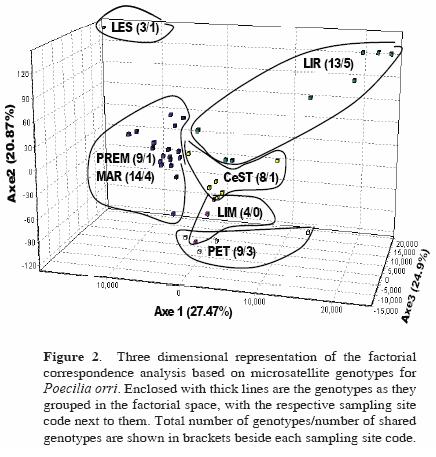
The factorial correspondence analysis based on microsatellite genotypes for P. orri (Fig. 2) showed the individuals' distinctive genotypes in each site, in which the 2 closest temporary wetlands (MAR and PREM) grouped closer together in the 3 dimensional representation. A permanent (LES) and a temporary (LIR) site had markedly distinctive genotypes compared to the rest of the populations. Finally, the remaining 2 permanent cenotes (CeST, LIM) showed a shared genetic composition, in proximity to a temporary wetland (PET). The temporary wetlands had the highest number of different genotypes (MAR = 14, LIR = 13, PREM = 9), while the permanent cenotes had the lowest number of genotypes (LES = 3, LIM = 4). Also, the most genotypically diverse sites were the ones that shared the highest number of genotypes among sites (LIR and MAR, with 5 and 4 shared genotypes respectively; see Fig. 2). On the other hand, no such clear resolution was observed for G. yucatana, in which only 1 site (LIR) showed a relatively isolated distribution (not shown).
Regarding the mitochondrial structure of P. orri, a 508 bp homologous fragment was recovered from the 24 individuals analyzed. No significant deviations from neutrality were noted (D = 0.746, p > 0.10; F = 1.299, p > 0.10). The number of polymorphic (segregating) sites observed was 26 (5.1%) and the total number of haplotypes was low (5): LES, LIM, CeST and LIR showed 1 haplotype, PET and MAR 2. Unique haplotypes were deposited in GenBank: accession numbers FJ769026–FJ769049. Accordingly, haplotype (h) and nucleotide diversity (π) estimates were low (h: range = 0–0.50; mean = 0.15; total sample = 0.64) and (π: range = 0–0.003; mean = 0.001; total sample = 0.022). The highest values were observed in PET (h = 0.50; π = 0.003) and MAR (h = 0.40; π = 0.001), while the rest had only 1 haplotype. Maximum parsimony searches generated 2 trees, with total lenght = 68, consistency index = 0.971 and retention index = 0.988. Two major groups (clades) were recovered as shown in the maximum parsimony tree (Fig. 3): Group 1 with haplotypes from CeST, LIM and PET, together with LES, and Group 2 that included haplotypes from LIR and MAR.
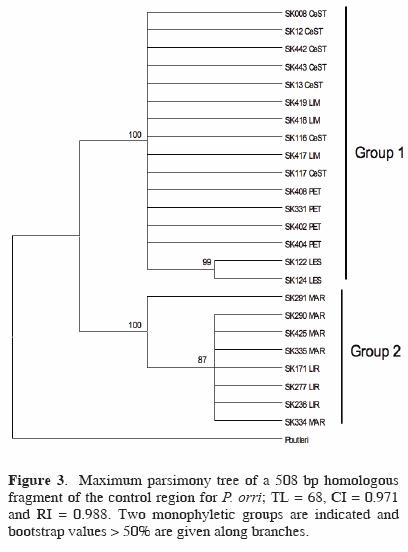
Values of nucleotide divergence (Dxy) were highest (4.3–4.7%) between the temporary wetlands LIR and MAR and also between the comparisons between both LIR and MAR and the other 4 water bodies (LES, LIM, CeST, PET), while the lowest values were amongst those 4 sites (0–0.3%), and between LIR and MAR (0.1%). AMOVA analyses indicated that genetic variability was significantly distributed among populations (92%) of the 6 water bodies sampled (ΦST = 0.918, p < 0.001). The divergence values observed between any of the populations of the water bodies and P. butleri —the species used as an outgroup— ranged from 10.7–10.9% (data not shown), while the divergence values observed between the 2 monophyletic groups found was 4.5% (see Fig. 3).
The minimum haplotype network (Fig. 4) showed that the most common haplotype (I), present in PET (3 individuals), CeST (7) and LIM (3) was separated by 1 mutational step from haplotype III (2 individuals from LES) and was separated by 3 mutational steps from haplotype II (1 from PET). The entirety of this latter group was separated by a high number of missing haplotypes (24) from the next most common haplotype (V), present in LIR (3) and MAR (4), separated this in turn by 2 mutational steps from an individual from MAR (haplotype IV). These results are congruent with the 2 groups depicted by the MP tree and by the divergence results (Figs. 3, 4).

Discussion
Besides the genetic study with isoenzymes and RAPDs with G. yucatana (Barona and Espinasa, 2004), our study represents the first analysis of genetic structure that includes 2 codistributed poeciliid species and compares their genetic diversity and structure, in both permanent and temporary water bodies (i.e. cenotes and wetlands), using microsatellites and mitochondrial DNA. Both fish species showed a relatively high number of alleles and of private alleles (microsatellites), but with levels of genetic diversity (heterozygostiy) markedly lower than expected and lower than average values reported for freshwater fishes (H0 = 0.540; DeWoody and Avise, 2000). This was more evident in P. orri, in which mitochondrial DNA (mtDNA) analyses also revealed low haplotype and nucleotide diversities. Low sample size in some sampling sites, mainly in LES and LIM, could be responsible for these low values, and we recognize that the present results must be interpreted with caution. Nonetheless, our results showed congruence between molecular markers (microsatellites and mtDNA) in P. orri, and provide the first description of genetic structure for this species that can serve as a basis for future studies.
The low intra–population variability observed with the loci studied is likely associated with the successive population size shrinkage and expansion events that occur on a seasonal–yearly basis in the wetlands of Sian Ka'an. Accordingly, little or none individual movement and gene flow, together with low population sizes and genetic bottlenecks, may be associated with low genetic diversity values. For example, populations of the endangered freshwater fish Ladigesocypris ghigii, endemic to the island of Rhodes, had high inter–population genetic structuring and extremely low levels of intra–population variability (1 or 2 haplotypes per population), regardless of a good sampling size (225 individuals and 9 populations). The authors interpret their findings as a result of successive bottleneck events related precisely to the shrinkage and expansion of populations inhabiting streams, springs and small reservoirs (Mamuris et al. 2005). Similarly, Grapputo et al. (2006) studied the effect of a severe bottleneck on genetic variability in 4 populations of the eastern mosquitofish (Gambusia holbrooki), and found a strong reduction of both the number of polymorphic loci and heterozygosity.
The cenote CeST had a very low heterozygosity value (0.026) despite having a sampling size above 10. Although cenotes do not shrink and expand, they are highly isolated, and CeST in particular is a special case, being the only water body inhabited by only 1 species of fish, P. orri, whereas the other 8 sites studied had a community composed of 3 to 11 different fish species (Zambrano et al., 2006). Despite the fact that it is a permanent cenote (i.e. it does not dry completely), the water level is significantly reduced during the dry season with a consequent high fish mortality likely associated with the low genetic diversity observed.
The genetic diversity values observed may also be related to the particular mating system that both species have: fertilization in poeciliids is internal and fertilized ova are retained within the ovary until the embryos are large enough to be self–sufficient and free swimming (Rosen and Bailey, 1963; Meffe and Snelson, 1989). They also have sperm storage and superfetation, i.e. successive brood overlap in the ovary of a single female (Chesser et al., 1984; Zane et al., 1999; Soucy and Travis, 2003). Such characteristics may allow these species to maintain a moderate level of genetic diversity via the introduction of different alleles into the populations, whereas the successive reduction of populations during dry periods results in diminished heterozygosity.
Populations that have more constant sizes and/or considerable genetic flow can maintain moderate to high levels of genetic diversity, even if they have small population sizes. The higher genetic variability observed for G. yucatana, with more than twice the heterozygosity levels of P. orri, jointly with the absence of genetic structure, is likely a result of this species having a better capacity for movement (i.e. gene flow). The latter because of its more euryhaline nature and its ability to sustain extreme environments (e.g. it withstands 0–37% salinity; Meffe and Snelson, 1989). For example, consider the biological characteristics of G. yucatana and the hydrology of the temporary wetland PET. PET has in its centre a tiny cenote that maintains some water when the entire wetland dries out, functioning as a refuge for some fish species. During the dry season we always found significantly higher numbers of G. yucatana in PET (and in most of the other water bodies; Zambrano et al., 2006), a time when some limnetic parameters attain extreme values, namely salinity, turbidity and temperature. G. yucatana is able to maintain individuals that can recolonize the wetland when it floods the following rainy season.
Different analyzes showed a consistent pattern: P. orri populations were found to be significantly structured and differentiated, compared to G. yucatana, which showed no genetic structure. Also, wetlands closer to each other consistently had lower genetic differentiation and divergence and higher number of migrants in P. orri than more isolated sites like some cenotes. Agreement was found on the basis of mtDNA data as well, given that nucleotide divergence was highest between the temporary and the permanent water bodies. Instead, in G. yucatana, low as well as high genetic distances were observed between both close and distant sites, regardless of whether or not they were cenotes or wetlands. These results agree with the acknowledgment that the outcome of frequent bottlenecks and/or extinction and recolonization events will be a combination of founder effects and genetic drift, which will enhance genetic differentiation among local populations (Slatkin, 1977; Whitlock and McCauley, 1990; Frankham et al., 2002). The study of Soucy and Travis (2003) exemplifies the above: they analyzed 7 populations of the poeciliid fish Heterandria formosa inhabiting rivers, lakes and ponds, and which exhibited significant variance in their histories of population density. The authors found, using 3 microsatellite loci as in the present study, that populations were genetically distinct and that reduced genetic diversity was found in populations with less density. Such distinctive genetic composition and differentiation was clearly illustrated in our study by the factorial correspondence analysis for P. orri, in which the 2 closest temporary wetlands MAR and PREM appeared as 1 group in the 3 dimensional representation (Fig. 2), indicating their unique genotypes. Temporary wetlands generally had the highest number of different genotypes, that is, they showed a higher genotypic diversity.
In a recent study that evaluates how habitat characteristics shape genetic variation in wild populations, Brouat et al. (2004) found that 5 environmental variables had a significant contribution in structuring inter and intrapopulational genetic diversity, based on canonical correspondence analysis and measured with microsatellite (neutral) markers. Our results of CCA showed that depth could be significantly associated with the population genetic structure in P. orri. Salinity, oxygen concentration and turbidity were important for G. yucatana, although they explained almost nothing of the total variation (3%), in accordance with the absence of genetic differentiation in this species.
The genetic structure of P. orri was likely a result of the intolerance of this species to extreme environments (e.g. high salinity and low oxygen concentration) and its biological characteristics (e.g. body size, reproductive attributes, and maturation age). P. orri individuals mature very rapidly in order to reproduce fast enough for the juveniles to reach maturity during the rainy season, whereas very few adults survive during the dry season (Meffe and Snelson, 1989). Survival of P. orri is therefore lower during dry periods, even in refuges like PET, where G. yucatana significantly outnumbered P. orri during our study (23 versus 5 individuals, Zambrano et al., 2006).
Specifically for P. orri, results of the parsimony analysis depicted 2 monophyletic groups, formed one by the cenotes LIM, CeST and LES, together with the wetland PET, and the other by the wetlands LIR and MAR, groups that showed a markedly high nucleotide divergence (4.5%). This genetic structure is supported by the minimum spanning network that resulted in 2 very divergent groups, which is indicative of a considerable long time of separation between these groups of water bodies (Templeton, 2004; Marcus et al., 2006). In addition, the cenotes group included the most common haplotype (i.e. the most probable ancestral haplotype; Clement et al., 2000). These results suggest that there is no accessible connection for the fish species studied and between the permanent cenotes and the wetlands evaluated, but further studies are needed to verify this observation.
The monophyly of the 2 groups and their relatively deep divergence (4.5%) is considerably high, especially considering that divergence values between P. orri and P. gilli (a phylogenetically close species to P. orri; Ptacek and Breden, 1998) is 3.3–4.6% (data not shown). Such genetic divergence value suggests that these lineages may be regarded as evolutionarily independent units. Mateos (2005) found a similar divergence (4.9%) between northern and southern populations of Poecilia butleri and suggested the possibility of 2 different species. Barona and Espinosa (2004) also found strong genetic and morphological differences between G. yucatana populations from 4 cenotes, and they suggest that the 4 populations could be regarded as different subspecies or even species. Undoubtedly, a wider study in terms of number of individuals and number of loci, together with a thorough morphological analysis, is needed to confirm this.
Our results illustrate that the isolation between temporary wetlands and permanent cenotes in Sian Ka'an, in combination with the successive flooding and drying cycles of these wetlands that cause the shrinkage and expansion of the P. orri and G. yucatana populations, likely have resulted in the low intrapopulation genetic variability observed and the markedly high genetic structuring of P. orri. Furthermore, our results show how 2 codistributed species, inhabiting the same hydrological system of cenotes and wetlands, can have a markedly different genetic structure, most probably determined by their particular biological and ecological characteristics. Further studies should be designed to evaluate in more detail the intricate relationship between environmental, biological and genetic attributes in these species. We also suggest that the assessment of abiotic characteristics in combination with the knowledge of biological attributes of species, evaluated through the view of their genetics, is a framework that can have valuable applications in ecological studies, and in conservation and management.
Acknowledgments
We thank D. García, G. Rodríguez, R. Vega, H.M. Castrejón and T. Camargo for their help in fieldwork. D. García helped with the identification of species. Amigos de Sian Ka'an and Federal Office of Sian Ka'an Biosphere Reserve provided lodging and support at the Santa Teresa field station. We thank the staff from the Sian Ka'an field station for their assistance. The project had financial support from Conacyt to EVD (139227V), from Semarnat–Conacyt to LZ (2002–01–0082), from Papiit (IN230007) to LZ, and from the Texas A & M–Conacyt program to LZ and EVD. Our study benefited enormously with the discussions and advise from William Loftus, Joel Trexler, Daniel Piñero and the population genetics and phylogeography discussion group at the Instituto de Ecología. Two anonymous reviewers made useful observations that helped improve the manuscript.
Literature cited
Barona, A. and L. Espinasa. 2004. Speciation in aquatic Trogloxenes in cenotes. AMCS Activities Newsletter 27:60–63. [ Links ]
Brouat, C., H. Chevallier, S. Meusnier, T. Noblecourt and J. Y. Rasplus. 2004. Specialization and habitat: spatial and environmental effects on abundance and genetic diversity of forest generalist and specialist Carabus species. Molecular Ecology 13:1815–1826. [ Links ]
Chesser, R. K., M. W. Smith and M. H. Smith. 1984. Biochemical genetics of mosquitofish III. Incidence and significance of multiple insemination. Genetica 64:77–81. [ Links ]
Clement, M., D. Posada and K. A. Crandall. 2000. TCS: a computer program to estimate gene genealogies. Molecular Ecology 9:1657–1660. [ Links ]
DeWoody, J. A. and J. C. Avise. 2000. Microsatellite variation in marine, freshwater and anadromous fishes compared with other animals. Journal of Fish Biology 56:461–473. [ Links ]
Excoffier, L., P. E. Smouse and J. M. Quattro. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491. [ Links ]
Excoffier, L., G. Laval and S. Schneider. 2005. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online 1:47–50. [ Links ]
Frankham, R., J. D. Ballou and D. A. Briscoe. 2002. Introduction to conservation genetics. Cambridge University Press, Cambridge, UK. [ Links ]
Fu, Y. X. and W. H. Li. 1993. Statistical tests of neutrality of mutations. Genetics 133:693–709. [ Links ]
Grapputo, A., A. Bisazza and A. Pilastro. 2006. Invasion success despite reduction of genetic diversity in the European populations of eastern mosquitofish (Gambusia holbrooki). Italian Journal of Zoology 73:67–73. [ Links ]
Hall, F. G. 1936. Physical and chemical survey of cenotes in Yucatán. In The cenotes of Yucatán, A. S. Pearse, A. P. Creaser and F. G. Hall (eds.) Carnegie Institute of Washington, Washington, D.C. p. 5–16. [ Links ]
Kocher, T. D., W. K. Thomas, A. Meyer, S. V. Edwards, S. Paabo, F. X. Villablanca and A. C. Wilson. 1989. Dynamics of mitochondrial DNA sequence evolution in animals. Proceedings of the National Academy of Sciences USA 86:6196–6200. [ Links ]
Levene, H. 1949. On a matching problem in genetics. The Annals of Mathematics Statistics 20:91–94. [ Links ]
Loew, S. S., D. F. Williams, K. Ralls, K. Pilgrim and R. C. Fleisher. 2005. Population structure and genetic variation in the endangered Giant Kangaroo Rat (Dipodomys ingens). Conservation Genetics 6:495–510. [ Links ]
Mamuris, Z., M. TH. Stoumboudi, C. Stamatis, R. Barbierai and K. A. Moutou. 2005. Genetic variation in populations of the endangered fish Ladigesocypris ghigii and its implications for conservation. Freshwater Biology 50:1441–1453. [ Links ]
Marcus, A. K., C. Kiefer, D. Ehrich, J. Vogel, C. Brochmann and K. Mummenhoff. 2006. Three times out of Asia Minor: the phylogeography of Arabis alpina L. (Brassicaceae). Molecular Ecology 15:825–839. [ Links ]
Mateos, M. 2005. Comparative phylogeography of livebearing fishes in the genera Poeciliopsis and Poecilia (Poeciliidae: Cyprinodontiformes) in Central Mexico. Journal of Biogeography 32:775–780. [ Links ]
Matoq, M. D., J. L. Patton and M. N. F. da Silva. 2000. Population genetic structure of two ecologically distinct Amazonian spiny rats: separating history and current ecology. Evolution 54:1423–1432. [ Links ]
Mcelroy, T. C., K. L. Kandl, J. García and J. C. Trexler. 2003. Extinction–colonization dynamics structure genetic variation of spotted sunfish (Lepomis punctatus) in the Florida Evergaldes. Molecular Ecology 12:355–368. [ Links ]
Meffe, G. K. and F. F. Jr. Snelson. 1989. Ecology and evolution of livebearing fishes (Poeciliidae). Prentice Hall. Englewood Cliffs. 453 p. [ Links ]
Meyer, A., T. D. Kocher, P. Basasibwaki and A. C. Wilson. 1990. Monophyletic origin of Lake Victoria cichlid fishes suggested by mitochondrial DNA sequences. Nature 347:550–553. [ Links ]
Nei, M. 1973. Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences of the USA 70:3321–3323. [ Links ]
Nei, M. 1987. Molecular evolutionary genetics. Columbia University Press, New York. 512 p. [ Links ]
Ostergaard, S., M. M. Hansen, V. Loeschcke and E. E. Nielsen. 2003. Long–term temporal changes of genetic composition in brown trout (Salmo trutta L.) populations inhabiting an unstable environment. Molecular Ecology 12:3123–3135. [ Links ]
Ptacek, M. B. and F. Breden. 1998. Phylogenetic relationships among the mollies (Poeciliidae: Poecilia: Mollienesia group) based on mitochondrial DNA sequences. Journal of Fish Biology 53 (Suppl. A):64–81. [ Links ]
Rice, W. R. 1989. Analyzing tables of statistical tests. Evolution 43:223–225. [ Links ]
Rodríguez, P. and H. Pizarro. 2007. Phytoplankton productivity in a highly colored shallow lake of a South American floodplain. Wetlands 27:1153–1160. [ Links ]
Rosen, D. E. and R. M. Bailey. 1963. The poeciliid fishes (Cyprinodontiformes), their structure, zoogeography, and systematics. Bulletin of the American Museum of Natural History 126:1–176. [ Links ]
Rozas, J., J. C. Sánchez–DelBarrio, X. Messeguer and R. Rozas. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497. [ Links ]
Sambroock, J., E. F. Fritswich and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York. 530 p. [ Links ]
Schmitter–Soto, J. J. 1999. Distribution of continental fishes in northern Quintana Roo, Mexico. Southwestern Naturalist 44:166–172. [ Links ]
Schmitter–Soto, J. J. and H. C. Gamboa–Pérez. 1996. Composición y distribución de peces continentales en el sur de Quintana Roo, península de Yucatán, México. Revista de Biología Tropical 44:199–212. [ Links ]
Schmitter–Soto, J. J., F. A. Comin, E. Escobar, J. Jerrera–Silveira, J. Alcocer, E. Suárez–Morales, M. Elías–Gutierrez, V. Díaz–Arce, L. E. Marín and B. Steinich. 2002. Hydrogeochemical and biological characteristics of cenotes in the Yucatan Peninsula (SE Mexico). Hydrobiologia 467:215–228. [ Links ]
Slatkin, M. 1977. Gene flow and genetic drift in a species subject to frequent local extinctions. Theoretical Population Biology 12:253–262. [ Links ]
Slatkin, M. 1987. Gene flow and the geographic structure of natural populations. Science 236:787–792. [ Links ]
Slatkin, M. 1995. A measure of population subdivision based on microsatellite allele frequencies. Genetics 139:457–462. [ Links ]
Snyder, J. R., A. Herndon and W. B. Robertson. 1990. South Florida Rockland. In Ecosystems of Florida. R. L. Myers and J. J. Ewel (eds.). University of Central Florida Press, Orlando. p. 230–276. [ Links ]
Soucy, S. and J. Travis. 2003. Multiple paternity and population genetic structure in natural populations of the poeciliid fish, Heterandria formosa. Journal of Evolutionary Biology 16:1328–1336. [ Links ]
Spencer, C. C., C. A. Chlan, J. E. Neigel, K. T. Scribner, M. C. Wooten and P. L. Leberg. 1999. Polymorphic microsatellite markers in the western mosquitofish, Gambusia affinis. Molecular Ecology 8:157–168. [ Links ]
Swofford, D. L. 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer Associates, Sunderland, Massachusetts. [ Links ]
Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin and D. G. Higgins. 1997. The Clustal–X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 24:4876–4882. [ Links ]
Templeton, A. R. 2004. Statistical phylogeography: methods of evaluating and minimizing inference errors. Molecular Ecology 13:789–809. [ Links ]
Trexler, J. C., W. F. Loftus and S. A. Perry. 2005. Disturbance frequency and community structure in a twenty–five year intervention study. Oecologia 145:140–152. [ Links ]
Vázquez–Domínguez, E., D. Piñero and G. Ceballos. 1999. Linking heterozygosity, demography, and fitness of tropical populations of Liomys pictus. Journal of Mammalogy 80:810–822. [ Links ]
Vázquez–Domínguez, E., G. Ceballos and D. Piñero. 2002. Exploring the relation between genetic structure and habitat heterogeneity in the rodent Liomys pictus from Chamela, Jalisco. Acta Zoológica Mexicana (n.s.) 86:17–29. [ Links ]
Whitlock, M. C. and D. E. McCauley. 1990. Some population genetic consequences of colony formation and extinction: genetic correlations within founding groups. Evolution 44:1717–1724. [ Links ]
Wright, S. 1931. Evolution in Mendelian populations. Genetics 10:97–159. [ Links ]
Yeh, F. C., R. C. Young, B. Timothy, T. B. J. Boyle, Z. H. Ye and J. X. Mao. 1997. PopGene, the user–friendly shareware for population genetic analysis. Molecular Biology and Biotechnology Center, University of Alberta, Alberta, Canada. (http//www.alberta.ca/). [ Links ]
Young, A. G. and G. M. Clarke. 2000. Genetics, demography and viability of fragmented populations. Cambridge University Press, Cambridge. 438 p. [ Links ]
Zambrano, L., E. Vázquez–Domínguez, D. García–Bedoya, W. F. Loftus and J. C. Trexler. 2006. Fish community structure in freshwater karstic wetlands of the Yucatán peninsula, Mexico. Ichthyological Exploration of Freshwaters 17:193–206. [ Links ]
Zane, L., W. S. Nelson, A. G. Jones and J. C. Avise. 1999. Microsatellite assessment of multiple paternity in natural populations of live–bearing fish, Gambusia holbrooki. Journal of Evolutionary Biology 12:61–69. [ Links ]














