Serviços Personalizados
Journal
Artigo
Indicadores
Links relacionados
-
 Similares em
SciELO
Similares em
SciELO
Compartilhar
Revista mexicana de biodiversidad
versão On-line ISSN 2007-8706versão impressa ISSN 1870-3453
Rev. Mex. Biodiv. vol.76 no.1 México Jun. 2005
Biogeografía
Historical biogeography in the age of complexity: expansion and integration
Biogeografía histórica en la era de la complejidad: expansión e integración
Daniel R. Brooks
Centre for Comparative Biology and Biodiversity, Department of Zoology, University of Toronto, Toronto, Ontario, Canada M5S 3G5
*correspondencia: dbrooks@zoo.utoronto.ca
Recibido: 27 de febrero del 2005
Aceptado: 11 de mayo del 2005
Abstract. Historical biogeography has recently experienced a significant advancement in three integrated areas. The first is the adoption of an ontology of complexity, replacing the traditional ontology of simplicity, or a priori parsimony; simple and elegant models of the biosphere are not sufficient for explaining the geographical context of the origin of species and their post-speciation movements, producing evolutionary radiations and complex multi-species biotas. The second is the development of a powerful method for producing area cladograms from complex data, especially cases of reticulated area relationships, without loss of information. That method, called Phylogenetic Analysis for Comparing trees (PACT), is described herein. The third element is the replacement of the model of maximum vicariance with the model called the Taxon Pulse hypothesis. Using PACT analysis for a data set of 33 different clades occurring in 9 different areas of endemism in Mexico, I show how taxon pulses can be detected. Finally, I show how PACT results can be used to provide a phylogenetic context for analyses of species-area relationships.
Key words: Phylogenetic analysis for comparing trees, PACT, taxon pulse hypothesis, areas of endemism, Mexico.
Resumen. Recientemente, la biogeografía histórica ha experimentado un avance significativo en tres aspectos integrales. El primero, es la adopción de una ontología de la complejidad, que reemplaza a la tradicional ontología de la simplicidad o parsimonia a priori; los modelos elegantes y sencillos para representar a la biósfera no son suficientes para explicar el contexto geográfico del origen de las especies y sus movimientos posteriores, generadores de radiaciones evolutivas y biotas multiespecíficas complejas. El segundo es el desarrollo de un método capaz de producir cladogramas de área a partir de datos complejos, especialmente casos de relaciones reticuladas de áreas, sin pérdida de información. Aquí describo ese método, llamado Análisis Filogenético para la Comparación de Árboles (PACT por sus siglas en inglés). El tercer aspecto, es la sustitución del modelo de máxima vicarianza por el modelo llamado hipótesis de pulsación de los taxa. Utilizando PACT para analizar 33 clados diferentes que ocurren en 9 áreas de endemismo en México, muestro cómo pueden detectarse las pulsaciones de los taxa. Finalmente, muestro cómo pueden utilizarse los resultados de PACT para proveer un contexto filogenético para el análisis de relaciones especies- área.
Palabras clave: Análisis filogenético para comparar árboles, PACT, hipótesis de pulsación de los taxa, áreas de endemismo, México.
Introduction
In a universe structured by laws, science is the search for theories providing powerful general explanations, and development of methods to explain data in terms of the general laws. This is the ontology of simplicity. Embodied in the principle of parsimony (Latin parcere, to spare), also known as the principle of simplicity. Aristotle (350 B.C.E.) postulated that «nature operates in the shortest way possible» and «the more limited, if adequate, is always preferable». This sense of the principle postulates that nature itself is parsimonious in some manner, and the principle is therefore ontological rather than epistemological. The principle is also linked with the English philosopher and Franciscan monk William of Ockham (ca. 1285-1349), who advocated the use of what is known as ‘Ockham‘s razor‘: « Pluralitas non est ponenda sine neccesitate » («plurality should not be posited without necessity») and « non sunt multiplicanda entia praeter necessitatem » («entities should not be multiplied unnecessarily»). In this sense, the principle of simplicity represents only an epistemological tool, or rule of thumb, which obliges us to favor theories or hypotheses that make the fewest unwarranted, or ad hoc, assumptions about the data from which they are derived. This epistemological use of parsimony does not necessarily imply that nature itself is parsimonious. Indeed, despite the best efforts of philosophers for more than 700 years, no link between parsimony and truth has ever been established. Nonetheless, most scientists conduct their research as if they believe that Nature is parsimonious in some sense, and they rely on theories that are simple.
In the second half of the 20 th century, historical biogeography produced two simple and elegant theories. The first of these was the Equilibrium Theory of Island Biogeography (ETIB) (MacArthur and Wilson, 1963; 1967). This theory is based on the view that dispersal from source areas to «islands» (actual or metaphorical), mediated by island size and distance, produces linear log-normal species-area relationships. Noise in the system, or the effects of contingency comprise in situ speciation and extinction. From this, one infers that data that conflict with the expected pattern (the «law») are the result of historical contingencies, and it is therefore permissible to remove or modify them. As a result, island biogeographers are admonished to study small, young islands, in order to minimize the potential for such historical contingencies that cloud our ability to see the true (and simple) pattern. The second simplicity theory was the Maximum Vicariance Hypothesis (MaxVic), also known as vicariance biogeography or cladistic biogeography (Humphries and Parenti, 1999). In contradistinction to ETIB, MaxVic is based on the theory that in situ speciation and extinction produce simple area cladograms in which each area appears once. Noise in the system, or the effects of contingency, result from dispersal. From this, one infers that data that conflict with a single area cladogram in which each area appears once (the «law») are the result of historical contingencies, and it is therefore permissible to remove or modify them. As a result, cladistic biogeographers developed Assumption 1 and 2 to remove or modify («reconcile») incongruent data with a single simple area cladogram.
One persistent concern about both these paradigms is this: If it is necessary to remove and modify data, and restrict one‘s scope of analysis, just how general and powerful are the explanations produced? A closer comparison of the two paradigms reveals another interesting feature: they are complementary theories, each one excluding the other‘s domain of explanation. What is missing from ETIB are assessments of the geographic origin of species, even though this is what distinguishes a «source» from an «island». What is missing in MaxVic are assessments of post-speciation movements, and yet this is how ancestral species become widespread enough to be affected by vicariance. These are not new observations - the original formulation of ETIB contained a term, g, meant to represent in situ phenomena, and early discussions of MaxVic acknowledged the importance of dispersal.
If each of these theories describes something valid, and each one excludes the other‘s explanatory domain, perhaps the problem lies in the adoption of ontological parsimony. That is, perhaps we should abandon simple theories. This requires the integration of three elements; (1) a formal basis for an ontology of complexity in evolution; (2) a method for detecting complexity in historical biogeographic relationships; and (3) a new model of biogeography that integrates both ETIB and MaxVic. I suggest that all three elements exist, and thus the basis for a new synthesis in historical biogeography is emerging.
1. The Ontology of Complexity in Evolution.
I submit that the ontology of complexity already exists - it is called Darwinism. Many forget that Darwinism is at its core not a simple theory. In Darwin ‘s own words
«... there are two factors: namely, the nature of the organism and the nature of the conditions. The former seems to be much more the important; for nearly similar variations sometimes arise under, as far as we can judge, dissimilar conditions; and, on the other hand, dissimilar variations arise under conditions which appear to be nearly uniform.»
-C. Darwin, 1872
a sentiment that was underscored more than 70 years later by one of the founders of the New Synthesis
«...in every part of the whole, wonderful history of life, all the modes and all the factors of evolution are inextricably interwoven. The total process cannot be made simple, but it can be analyzed in part. It is not understood in all its appalling intricacy, but some understanding is in our grasp, and we may trust our own powers to obtain more.»
- G.G. Simpson, 1944
Darwin used two metaphors, a phylogenetic tree and the tangled bank, to visualize the complexity of evolution. By referring to species as «communities of descent», Darwin emphasized that the fundamental explanatory principle is shared history. Evolution has been so complex and historically contingent, however, that the history includes both general (lawlike) and unique (contingent) phenomena. Extending this to biogeography leads us to predict that historical biogeographical patterns should be historically unique combinations of dispersal (ETIB) and in situ events (MaxVic). Furthermore, we would predict that our ability to document those patterns would be obscured most by the use of models and methods that over-simplify the process by invoking a priori assumptions or prohibitions. This leads us to recognize several essential elements of the analytical method required to study historical biogeography as a complex phenomenon.
First, it is not permissible to remove or modify data. Wiley (1986, 1988a,b) and Zandee and Roos (1987) already formalized this as «Assumption 0,» which states that you must analyze all species and all distributions in each input phylogeny without modification, and your final analysis must be logically consistent with all input data. Recognition of the fundamental importance of Assumption 0 was obscured by Page (1990), who used «Assumption 0» to refer to the protocol of coding «absence» as «0» in preparing a matrix of data for analysis. Brooks (1981) proposed that protocol because computer programs at that time did not accept missing data. It was eliminated when Wiley (1986) proposed using missing data coding for absences for analyses using Brooks‘ method, which Wiley dubbed Brooks Parsimony Analysis (BPA). The confusion over what was really Assumption 0 led Van Soest and Hajdu (1997) to propose what they called a new NA (no assumption) protocol, apparently not realizing this was BPA a la Wiley (1986). Even more recently, Porzecanski and Cracraft (2005) proposed modifications of Parsimony Analysis of Endemicity (PAE) apparently not realizing they were also reinventing BPA a la Wiley (1986).
Second, complex area cladograms must include reticulated area relationships. If each area on this planet had a singular history with respect to all the species living in it, either there would be either one species per area or one clade per area. Nowhere on earth does this occur, so we must assume that reticulated area relationships have been common. If we use a method of analysis that produces simple area cladograms (i.e. ones in which each area appears only once), Assumption 0 will be violated whenever an area has a reticulated history. Assumption 0 can be satisfied in such cases by duplicating areas with reticulated histories. Therefore, a method of analysis for handling complexity requires a Duplication Rule, a mechanism by which areas are listed for each evolutionary event affecting them.
Finally, if you allow all possibilities, including area reticulations, a priori for all species in each clade being analyzed, and if we expect historical biogeographical patterns to be combinations of unique and general phenomena, how can you find the general patterns? For this, we use an epistemological corollary of the Duplication Rule - Make only enough duplications to satisfy Assumption 0. This is simply a rendering of Ockham‘s Razor - Do not duplicate areas beyond necessity. Simplicity is thus used to determine IF there are general patterns, it is not used to impose simplicity on the data.
2. A Method for Detecting Complex Historical Biogeographical Patterns.
Recent studies of the general properties of the methods of cladistic biogeography have shown that all of them behave in internally inconsistent ways when dealing with complex data (Van Veller et al., 1999, 2000, 2001, 2002, 2003; Dowling, 2002; Dowling et al., 2003). In choosing a parsimonious model of biotic evolution, advocates of vicariance biogeography, and particularly cladistic biogeography, have developed methods that explained geographic distributions of sister groups based on a restricted range of evolutionary processes. All methods recognize three classes of biogeographic patterns: (1) complete matching between the general pattern and any given taxon-area cladogram, usually interpreted as indicating vicariance, but recognized by some as possibly being the result of sequential speciation by colonization in each clade (Fig. 1); (2) incomplete matching, suggesting extinction in one of the lineages (also known as «lineage sorting») (Fig. 2); (3) duplication of all or part of the pattern, suggesting sympatric speciation in the common ancestor of the duplicated lineages (also known as «lineage duplication») (Fig. 3).

Figure 1. A taxon-area cladogram showing a particular set of area relationships involving areas A, B, C, and D, stipulated to be the general pattern (left), and a second taxon-area cladogram showing the same area relationships as the general pattern (right). Letters = areas.

Figure 2. A taxon-area cladogram showing a particular set of area relationships involving areas A, B, C, and D, stipulated to be the general pattern (left), and a second taxon-area cladogram showing area relationships among areas A, C, and D, interpreted as having lost, through extinction (also known as lineage sorting) a species occurring in area B that was the sister species of the common ancestor of the species occurring in areas C and D (right). Letters = areas.
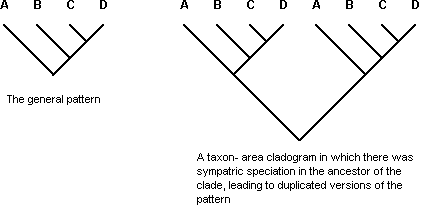
Figure 3. A taxon-area cladogram showing a particular set of area relationships involving areas A, B, C, and D, stipulated to be the general pattern (left), and a second taxon-area cladogram of two major parts, each of which shows the same area relationships as the first taxon-area cladogram, interpreted as having experienced a sympatric speciation event (lineage duplication) in the common ancestor of the clade (right). Letters = areas.
Three additional types of patterns have been considered complicating factors that obscure the general area relationships, and have been excluded from the simple area cladograms produced by all methods of vicariance biogeography except the one known as secondary BPA (Brooks, 1990; Brooks and McLennan, 1991, 2002; Brooks et al., 2001). One of these is speciation by dispersal on the part of one or more members of the co-occurring clades (peripheral isolates allopatric speciation), introducing unique area relationships (Fig. 4). The remaining two types of patterns represent cases in which more than one phylogenetic event affects the same area, producing reticulated area relationships: (a) two or more separate speciation events within a clade each resulting in at least two non-sister species inhabiting the same area (Fig. 5) and (b) post-speciation dispersal leading to the occurrence of the same species in more than one area (also known as the widespread species problem) (Fig. 6). If the geography of evolution has been complex, the methods of vicariance biogeography will produce internally inconsistent results in direct proportion to the complexity in real data the methods must explain away with auxiliary assumptions, even if they are called costs, likelihoods, or probabilities. Or, (ontological) simplicity is not always the most parsimonious (epistemological) depiction of the real world (Van Veller and Brooks, 2001).
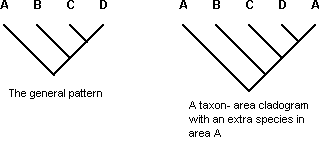
Figure 4. A taxon-area cladogram showing a particular set of area relationships involving areas A, B, C, and D, stipulated to be the general pattern (left), and a second taxon-area cladogram showing area relationships among areas A,B,C,D, and E, with the addition in area E of a sister species of the species occurring in area B in the second taxon-area cladogram, interpreted as an instance of peripheral isolates speciation (allopatric speciation by dispersal) (right). Letters = areas.

Figure 5. A taxon-area cladogram showing a particular set of area relationships involving areas A, B, C, and D, stipulated to be the general pattern (left), and a second taxon-area cladogram showing the same area relationships, with the addition of a species in area A that is the sister species of the species occurring in area D, indicating that the species occurring in area A arose from two different ancestors. Area A is thus said to have a reticulated history (right). Letters = areas.

Figure 6. A taxon-area cladogram showing a particular set of area relationships involving areas A, B, C, and D, stipulated to be the general pattern (left), and a second taxon-area cladogram showing the same area relationships as the first taxon-area cladogram, except that the species occurring in area A also occurs in area D; and a general area cladogram representing the area relationships supported by both taxon-area cladograms (right). The species occurring in areas A and D is interpreted as a case of post-speciation dispersal from area A to area D. Letters = areas.
Recent studies using secondary BPA have shown extensive dispersal and reticulated area relationships (Spironello and Brooks, 2003; Bouchard et al., 2004), even for data sets carefully chosen to emphasize vicariance (e.g., Brooks and McLennan, 2001; McLennan and Brooks, 2002; Halas et al., 2005). Most notable among these was the discovery that 70% of the areas recognized in the so-called «classic case of vicariance,» the Mesoamerican freshwater fishes Xiphophorus and Heterandria, are not vicariant areas of endemism, and have reticulated biogeographical histories (Green et al., 2002).
As noted above, secondary BPA is the only method of vicariance biogeography that attempts to depict the full range of distributions exhibited by all species in multiple taxon-area cladograms, including widespread species and reticulated area relationships. BPA can be implemented using standard methods in phylogenetic analysis (Brooks and McLennan, 2002, 2003), but only with laborious manipulations of the data. All taxon-area cladograms need to be converted into binary matrices, and each area duplication requires that the matrix be re-formulated. This re-formulation produces large numbers of pseudo-missing data codes representing the areas not affected by the unique events requiring the duplication (Brooks and McLennan, 1991, 2002). Performing such an analysis for complex data sets is thus time-consuming. In addition, because one cannot specify the number and types of area duplications that will be needed a priori, some have been led to believe that the duplication convention in BPA idiosyncratic rather than algorithmic (e.g., Ronquist, 2002; Siddall and Perkins, 2003).
Wojcicki and Brooks (2005) produced an algorithm for deriving area cladograms that embodies the strengths of Secondary BPA while eliminating its weaknesses. The inspiration for this algorithm comes from considering Venn diagram representations of host cladograms as strings of hierarchically organized characters. The algorithm uses the string input to build a tree-like data structure that can be searched for points of agreement and disagreement with additional input host cladograms (Cormen et al., 2001). We assume that the history of the host context of speciation, dispersal, and extinction for any assemblage of parasite clades comprises a long and complex combination of strings. We also assume that no single parasite clade contains the complete information, even about its own particular history. By combining the partial information from each of many parasite clades, however, we can reconstruct substantial parts of the coevolutionary record of life by integrating information from multiple clades. Since the hierarchical organization of the strings of characters stems from phylogenetic relationships, we refer to this algorithm as Phylogenetic Analysis for Comparing Trees (PACT).
3. A «New» General Theory of Historical Biogeography: The Taxon Pulse.
Formal methods of historical biogeographic analysis using phylogenetic trees began appearing more than 25 years ago (Platnick and Nelson, 1978). At the time, their conceptual underpinnings seemed straightforward. Episodes of allopatric speciation resulting from the formation of a geographic barrier, called vicariant speciation, or vicariance, would produce biogeographical patterns of distributions of sister species mirroring the history of barrier formation. Furthermore, such barrier formation would affect multiple clades at the same time, so the biogeographic patterns produced by episodes of vicariance would be general, or redundant patterns. Phenomena such as postspeciation dispersal, peripheral isolates speciation (allopatric speciation by dispersal), and extinction, were assumed to be clade-specific, producing patterns incongruent with the general area relationships. The research program became widely known as vicariance biogeography following the publication of the proceedings of a major symposium (Nelson and Rosen, 1981) and of a text devoted to the subject (Nelson and Platnick, 1981).
Vicariance biogeography has spawned two distinct research programs. Cladistic biogeography (Humphries and Parenti, 1999) is based on the view that the function of historical biogeography is to determine general area relationships, and that each area has a singular history with respect to the species occurring in it; in a sense, to produce a phylogeny of areas. Phylogenetic biogeography sensu Van Veller and Brooks (2001; see also Brundin, 1966, 1972), by contrast, views historical biogeography as a means to assess the temporal and spatial context of evolutionary radiations, modes of initiating speciation, and sequences of biotic assembly (Brooks and McLennan, 1991, 2002). Despite their fundamentally different perspectives on the goals of historical biogeography, advocates of both programs have always agreed that general biogeographic patterns are the result of vicariance.
At nearly the same time the maximum vicariance paradigm emerged, Erwin (1979, 1981) proposed the taxon pulse hypothesis as a model incorporating both dispersal and vicariance. Erwin‘s model stemmed from an idea proposed by Darlington (1943) later named the «taxon cycle» by Wilson (1959, 1961). Taxon pulse and taxon cycle models both assume that species and their adaptations arise in «centers of diversification» and that distributional ranges of taxa periodically fluctuate around a more stable, continuously occupied centre. This general biotic dispersal may be interrupted by the formation of barriers, producing episodes of vicariant speciation. Breakdown of those barriers produces new episodes of biotic expansion, setting the stage for yet more episodes of vicariance. Taxon cycles occur over relatively short periods of time («ecological time») and involve species that disperse actively and colonize new areas during expansion episodes, then contract their ranges during periods of habitat contraction, without producing new species. Taxon pulses, by contrast, occur over relatively long periods of time («evolutionary time») and are characterized by dispersal along a broad front during expansion into suitable habitat when previous barriers break down. During this expansion phase, different species within a biota encounter additional geographic heterogeneity, including range contractions. Such heterogeneity may: (1) stop the expansion of some species, resulting in species of restricted distributions; (2) affect only the rate of expansion for some species, producing widespread species; or (3) act as barriers to dispersal of sufficient magnitude to produce new species as a result of peripheral isolates speciation. Geological evolution, operating on longer time scales than biological evolution, may also produce barriers, resulting in episodes of vicariant speciation affecting members of these same biotas.
Despite the existence of an alternative to maximum vicariance, and despite concerns that exemplar taxa were being carefully selected to show a preponderance of vicariance (Simberloff et al., 1981; Simberloff, 1987), vicariance has become the default explanation for any observation of allopatry. And yet, the maximum vicariance model has always been deficient because it neglects the issue of how ancestral species of many clades become widespread enough to be affected by vicariant events. If vicariance affects many members of ancestral biotas in the same way, it seems reasonable to assume that at some point in the past, the members of the biota expanded their geographic ranges to such an extent that they could be affected by the subsequent vicariance event. Advocates of vicariance biogeography have acknowledged that this must happen: Wiley (1981) noted that some circumstances, such as colonization of islands, might produce general distribution patterns based on dispersal rather than vicariance, and Endler (1982) suggested that such correlated dispersal patterns might be common. In practice, however, historical biogeographers have simply assumed that such dispersal does not produce general patterns, so it is permissible to invoke dispersal only to explain departures from the general pattern, which is always explained as the result of vicariance (Wiley, 1986, 1988a,b; Brooks and McLennan, 1991, 2002).
Taxon pulse-driven biotic diversification differs from vicariance-driven biotic diversification in three important ways. First, because diversification is driven by biotic expansion, we expect to find general patterns associated with dispersal, not just with vicariance. General patterns resulting from biotic expansion occur when barriers to dispersal, especially the large-scale ones leading to vicariance, break down. Second, episodes of biotic expansion, even those involving large areas, will inevitably lead to reticulated historical relationships among areas, and biotas within areas of endemism comprising species of different ages derived from different sources. Third, the absence of particular clades in particular areas is more parsimoniously explained as a lack of participation in that particular expansion episode by a particular clade, rather than dispersal with extinction. Taxon pulses are also historically contingent, meaning that at any given time, different clades comprising a complex biota may form a mosaic of area relationships. Halas et al. (2005) illustrated their protocols using the extensive data set presented by Marshall and Liebherr (2000), representing 33 clades of insects, vertebrates, and flowering plants, occurring throughout Mexico and parts of Central America, which is particularly relevant for this contribution.
Materials and Methods
Producing the area cladogram. A Precis of PACT.
Step 1. Convert all phylogenetic trees of interest into taxon-area cladograms. This is accomplished by replacing the names of the species with the areas they inhabit.
Step 2. Convert the taxon-area cladograms into Venn diagrams (Table I). The Venn diagrams comprise two classes of elements, «leaves» and «nodes». A leaf is a single area, and a node is any grouping of at least 2 areas. Nodes are represented by inclusive open [«(«] and closed [«)»] parentheses in the Venn diagram. When a given species inhabits more than one area, a leaf designates each of the areas and all the areas inhabited by that species are contained within a single node.

Step 3. Choose any taxon-area cladogram from the set of taxon-area cladograms to be analyzed, and determine its elements. We will refer to this as the Template Area Cladogram.
Template Area Cladogram (Taxon-area Cladogram 1 in Table I): (A(B(CD)))
The algorithm reads the Venn diagram representing the second taxon-area cladogram from left to right, element by element. Each time a closed parenthesis [')'] is encountered, indicating a grouping of at least 2 areas, the algorithm moves backwards, until it reaches an open parenthesis [‘(‘ ], collecting the data for the grouping thus created. Next, the algorithm represents the grouping signified by the inclusive parentheses by a node, which is a data structure designating a grouping and which is used in integrating the taxon-area cladogram with the template area cladogram. In this case, the first closed parenthesis is reached after D. The algorithm then reads backwards (to the left) collecting leaves and nodes until it reaches the open parenthesis, in this case C + D. Once the data collection is complete a node containing the leaves CD replaces the parentheses around C and D. If we called that node «X», the Venn diagram would now be (A(BX)). The algorithm continues reading to the left, searching for the next open parenthesis. The next open parenthesis bracket forming a node, ‘Y‘, containing the leaf B and X. The taxon-area cladogram is now modified to (AY), a grouping that receives its own node, ‘Z‘. The template area cladogram is now represented by 4 leaves and 3 nodes: A, B,C, D, Z[A(B(CD)))], Y [(B(CD))], and X [(CD)].
Step 4. Select a second taxon-area cladogram. Determine its elements as in step 1, and then compare each of them with the template area cladogram.
Template Area Cladogram: (A(B(CD)))
Taxon-area cladogram 2 (Table I): (A(B(CD)))
PACT reads the second taxon-area cladogram in the same manner as it read the template area cladogram. In this case, the first closed parenthesis is reached after D. PACT then reads to the left, collecting leaves and nodes until it reaches the open parenthesis, in this case it collects leaves C + D. Once the data collection is complete, the parentheses around C and D are replaced by a node containing the leaves CD. The next open parenthesis forms a node, containing the leaf B and the node (CD). Finally, the last open parenthesis forms a node containing the leaf A and the node (B(CD)). The taxon-area cladogram is now represented by 4 leaves and 3 nodes: A, B,C, D, (A(B(CD))), (B(CD)), and (CD). The next step is to integrate the taxon-area cladogram with the template area cladogram. This is accomplished by maximizing the matches between their respective leaves and nodes, and then adding novel elements by creating novel nodes at appropriate levels in the template area cladogram. Next, PACT re-reads the elements of the taxon-area cladogram, comparing them with the elements of the template area cladogram. Each element in the input taxon-area cladogram that also occurs in the template area cladogram is designated with a ‘Y‘; any element of the input taxon-area cladogram that is not found in the template area cladogram is designated with a ‘N‘:
(A(B(CD))) -Y; A-Y + (B(CD))-Y; B-Y + (CD)-Y; C-Y + D-Y
This produces the first, and most basic rule of PACT, the ‘Y + Y = Y‘ rule. In this case, each element of the input taxon-area cladogram is congruent with an element in the template area cladogram (all elements in tree 2 are Y‘s), so trees 1 and 2 can be combined completely. The general area cladogram resulting from the combination of trees 1 and 2 is thus (A(B(CD))) (Fig. 7).

Figure 7. PACT-derived area cladogram for taxon-area cladograms 1- 4 in Table 1. Letters = areas.
PACT performs this search in the sequence in which groups appear in the input taxon-area cladogram to be combined with those in the template area cladogram. This speeds up the process of analyzing the new cladogram and making combinations and addition to the template. In the case above, for example, PACT would have recognized that, because (A(B(CD))) = Y in the input taxon-area cladogram, all elements in the input taxon-area cladogram corresponded to elements in the template area cladogram, and would have made the combination immediately.
Step 5. Add a third taxon-area cladogram (tree 3), and repeat steps 2 and 3, comparing it with the tree resulting from the combination of the previous steps.
Template Area Cladogram: (A(B(CD)))
(A(B(CD))); A + (B(CD)); B + (CD); C + D
Taxon-area Cladogram 3 (Table I): A(CD))
(A(CD))-N; A-Y + (CD)-Y; C-Y + D-Y
In this case, there is a mismatch between the template area cladogram and the input taxon-area cladogram at the initial level, indicated by N. At this point no decision can be made as to why the mismatch occurs, so PACT does not produce any changes and moves on. All remaining elements in taxon-area cladogram 3 are ‘Y‘, so we can combine them with the template area cladogram. At this point we can begin to consider the mismatch, but we discover that in this case, the entire input taxon-area cladogram has been combined with the template area cladogram. The ‘N‘ seems to have disappeared. The reason for this is that the template area cladogram differs from the input taxon-area cladogram only by containing information not found in the input taxon-area cladogram. The absence of B in the input taxon-area cladogram does not affect the placement of B in the template area cladogram, and thus does not affect the topology of the area cladogram. The general area cladogram for trees 1+2+3 is still (A(B(CD))) (Fig. 7).
Step 6. Add the next tree (4) and repeat steps 2-3.
Template Area Cladogram: (A(B(CD)))
(A(B(CD))); A + (B(CD)); B + (CD); C + D
Taxon-area Cladogram 4 (Table I): (A(B(CD)))(A(B(CD))))
(A(B(CD)))(A(B(CD))))-N; (A(B(CD)))-Y; A-Y + (B(CD))-Y; B-Y + (CD)-Y; C-Y + D-Y; (A(B(CD)))-Y; A-Y + (B(CD))-Y; B-Y + (CD)-Y; C-Y + D-Y
Once again, the only N occurs at the level of the entire input taxon-area cladogram, and that N disappears once the lower levels are combined with the template. In this case, the input taxon-area cladogram appears more complex than the template, but only because it contains two identical representations of the template area cladogram. This is the diagnostic signature of lineage duplication, sympatric speciation within an ancestor producing two co-occurring lineages. This does not affect the pattern of relationships among areas, so the general area cladogram for trees 1+2+3+4 is still (A(B(CD))) (Fig. 7).
Step 7. Add the next tree (5) and repeat steps 2-3.
Template Area Cladogram: (A(B(CD)))
(A(B(CD))); A + (B(CD)); B + (CD); C + D
Taxon-area Cladogram 5: (A((BE)(CD)))
(A((BE)(CD)))-N; A-Y + ((BE)(CD))-N; (BE)- N + (CD)-Y; B-Y + E-N; C-Y + D-Y
Reading from the left, PACT encounters (BE); B in the input taxon-area cladogram is Y, and because B and BE are connected at the same node, both B‘s can be combined. E, which is not found in the template area cladogram, is thus a novel (‘N‘) element, and added to the template area cladogram at that point, creating a (BE) grouping (a new node) in the template. The next closed parenthesis is encountered at (CD); both C and D as well as the grouping CD are Y in the template area cladogram, so there is no change at this point. The next closed parenthesis is ((BE)(CD)). This combination already exists in the template area cladogram due to the modification made earlier in which E was added to the template area cladogram. Finally, PACT encounters A, which is Y, and is combined with the template. The resulting area cladogram is (A((BE)(CD))) (Fig. 8).
Step 8. Add the next tree (6) and repeat steps 2-3, comparing it with the tree produced by 1+2+3+4+5 (Fig. 8).
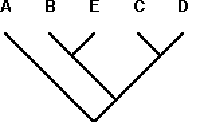
Figure 8. PACT-derived area cladogram for taxon-area cladograms 1- 5 in Table 1. Letters = areas.
Template Area Cladogram: (A((BE)(CD))))
(A((BE)(CD)))); A + ((BE)(CD)); (BE) + (CD); B + E; C + D
Taxon-area Cladogram 6: (A(B(C(DA))))
(A(B(C(DA))))-N; A-Y* + (B(C(DA)))-N; B-Y + (C(DA))-N; C-Y + (DA)-N; D-Y + A- Y*
This case is directly analogous to the previous one. Reading from left to right, PACT initially encounters (AD), which is not found in the template area cladogram (‘N‘). The ‘A‘ in (AD) is thus considered a novel element (‘N‘) and the input taxon-area cladogram is modified to
(A(B(C(DA))))-N; A-Y + (B(C(DA)))-N; B-Y + (C(DA))-N; C-Y + (DA)-N; D-Y + A-N
Next, PACT encounters (CD) in the template tree and (C(DA)) in the input taxon-area cladogram tree. C is a common element in both cladograms, and can be combined. This leaves D in the template area cladogram and (DA) in the input taxon-area cladogram connected at the same node. This means that both D‘s can be combined, creating a (C(DA)) grouping (and new node) in the template area cladogram. At the next node, we find the grouping (BE) in the template area cladogram and the leaf B in the input taxon-area cladogram. As in step 7, above, both B‘s can be combined, leaving the grouping (BE) in the template area cladogram inPACT. At the next level, we encounter leaf A in both cladograms, which are combined. This confirms PACT‘s initial assessment of Y for leaves A (basal most), B, C and D. The input taxon-area cladogram contains a novel grouping (DA) not found in the template and the template contains a grouping (BE) not seen in the input taxon-area cladogram. The resulting area cladogram is (A((BE)(C(DA)))) (Fig. 9).

Figure 9. PACT-derived area cladogram for taxon-area cladograms 1- 6 in Table 1. Letters = areas.
The situation presented by taxon-area cladograms 5 and 6, above, represent cases of what we call the ‘Y + YN = YN‘ rule. For clade 5, ‘Y‘ = B and ‘YN‘ = BE; for clade 6 ‘Y‘ = D and ‘YN‘ = DA. Next, we consider taxon-area cladograms 7 and 8 in Table I on their own, in order to demonstrate a final combination rule.
Step 9. Choose one area cladogram to be the template (we choose 7 in this case, but one could also choose 8 without changing the results).
Taxon-area Cladogram 7: (A(BE))
Taxon-area Cladogram 8: (A(CD))
(A(BE)); A + (BE); B + E (A(CD))-N; A-Y + (CD)-N; C-N + D-N
A is the only common element (Y) in both taxon-area cladograms. The groups (BE) and (CD) contain no elements in common, but each is connected at a node with A. In this case, although many dichotomous area cladograms consistent with the data are possible, we have no evidence supporting any particular one. Therefore, the resultant area cladogram is (A(BE)(CD)) (Fig. 10). This is an example of what we call the ‘YN + YN = YNN‘ rule, where ‘A‘ = Y, ‘(BE)‘ = N and ‘(CD)‘ = N.
Step 10. We can now combine the area cladogram for taxon-area cladograms 7 and 8 (Fig. 10) with the template area cladogram (Fig. 9).

Figure 10. PACT-derived area cladogram for taxon-area cladograms 7- 8 in Table 1. Letters = areas.
Template Area Cladogram for clades 1-6: (A((BE)(C(DA)))) Area cladogram for clades 7-8: (A(BE)(CD))
(A(BE)(CD))-N; A-Y + (BE)-Y + (CD)-N; B-Y + E-Y; C-Y + D-Y
D in the area cladogram for clades 7-8 and (DA) in the template area cladogram is a case of the ‘Y + YN = YN‘ rule, so D in the area cladogram for areas 7-8 is combined with D in (DA) in the template area cladogram); therefore (CD) in the area cladogram for clades 7-8 is combined with (C(DA)) in the template. A and (BE) are both Y, so they are combined; At this point, all elements in the area cladogram for clades 7-8 have been integrated with the template area cladogram (A((BE)(C(DA)))) (Fig. 9).
Step 11. Add taxon-area cladogram 9 in table I to the Template Area Cladogram.
Template Area Cladogram: (A((BE)(C(DA))))
(A((BE)(C(DA)))); A + ((BE)(C(DA))); (BE) + (C(DA)); C + (DA); B + E; D + A
Taxon-area Cladogram 9: (A(A(B(CD))))
(A(A(B(CD))))-N; A-Y* + A(B(CD))-N; A-Y* + (B(CD))-N; B-Y + (CD)-N; C-Y + D-Y
Once again, reading from left to right, the algorithm first encounters (CD). We begin with CD in the input taxon-area cladogram + (C(DA)) in the template area cladogram. This is a case of the ‘Y + YN = YN‘ rule, so D in the taxon-area cladogram is combined with (DA) in the template area cladogram. Next, (B(CD)) in the input taxon-area cladogram, now considered (B(C(DA))), is connected at the same node in the template area cladogram as ((BE)(C(DA))). (C(DA)) is the same in both cases, so they are combined, leaving B and (BE), another case of the ‘Y + YN = YN‘ rule. At the next node, the template area cladogram and the taxon-area cladogram are both A, so they are combined.
Finally, the input taxon-area cladogram has an additional A, originally designated Y, because A occurs twice in the template area cladogram and twice in the taxon-area cladogram. At this point, we have already accounted for both A‘s in the template area cladogram, so PACT must still account for the second A in the input taxon-area cladogram. One possibility is that the two A‘s in the taxon-area cladogram are paraphyletic because they represent an episode of sympatric speciation (lineage duplication), in which case both could be combined. PACT does not combine these A‘s, for a methodological and a biological reason, respectively. First, single areas are not sufficient grounds for grouping or combining areas. PACT will not create groupings of areas in the absence of any evidence of groupings. Second, sympatric speciation is not the only possible explanation for the paraphyletic status of the area A‘s. Combining the two A‘s would be tantamount to making a choice in favor of sympatric speciation in the face of ambiguity, rather than waiting for additional data (more taxon-area cladograms) to resolve the ambiguity.
This provision in PACT prevents over-combining data; we call it the «Y(Y-» = «Y(Y-»or «Y(Y- ¹ Y» rule, or «do not combine single common areas attached to different nodes». All current methods, including secondary BPA, violate this rule by over-combining data. In matrix representation methods, including BPA, this is called inclusive ORing, which is known to create other systemic analytical problems (Cressey et al., 1983; Brooks and McLennan, 1991, 2002) and internal inconsistencies (Van Veller et al., 1999, 2000, 2001, 2002). Consider the taxon-area cladograms ((AC)B) + (A(AB)). PACT produces (A((AC)B)) for these two taxon-area cladograms. If we combine the A‘s in taxon-area cladogram 2, the result would be ((AC)B). Now add a third taxon-area cladogram, (A(CB)). The PACT result is still (A((AC)B)), supporting an interpretation that all 3 taxon-area cladograms are parts of a single complex pattern, one part of which is missing in each. If we had combined the A‘s in taxon-area cladogram 2, however, the result would be an unresolved polytomy (ACB). At this point, all methods, including secondary BPA, would infer that the taxon-area cladograms had no information in common.
PACT thus treats the basal-most A as a new element added to the template area cladogram, which is modified to (A(A((BE)(C(DA)))))
(Fig. 11).
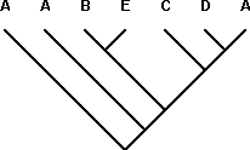
Figure 11. PACT-derived area cladogram for taxon-area cladograms 1- 9 in Table 1. Letters = areas.
All available taxon-area cladograms have now been incorporated, resulting in the final area cladogram (Fig. 11). Some may notice at this point that either of the two basal A‘s in taxon-area cladogram 9 could be considered the same as the basal A in the template area cladogram. This ambiguity does not affect the construction of the area cladogram, only the mapping of particular species onto the area cladogram when we begin to derive evolutionary inferences from the area cladogram.
Distinguishing General Nodes due to Vicariance from General Nodes due to Biotic Expansion.
Lieberman (2000, 2003a, b) proposed a protocol for distinguishing general nodes due to vicariance from those due to biotic expansion in area cladograms. General nodes associated with vicariance exhibit decreasing numbers of areas occupied, whereas general nodes associated with biotic expansion are associated with increasing numbers of areas occupied (break-down of a barrier). Vicariance nodes should also be characterized by splits between areas caused by the documented formation of a geological, geographical, or climatological barrier of sufficient magnitude and duration to produce speciation (i.e., irreversible splitting of lineages) in multiple clades. For ambiguous cases, we assume vicariance as the default explanation.
Results and Discussion
The core of the PACT area cladogram for the Marshall and Liebherr (2000) data set is shown in Fig. 12. The remaining portions, which can be obtained from the author, depict a complex biogeographic history in which every area shows evidence of reticulation. The area cladogram in Fig. 12 has 15 nodes (Roman numerals). Following Lieberman‘s rationale, six of those nodes are vicariance nodes. Three correspond to repeated episodes of the same split, between the Transmexican Volcanic Belt + Sierra Madre del Sur and the Chiapan Guatemalan Highlands + Talamancan Cordillera: Node I, represented by 7 clades; Node VIII, represented by 21 clades; and Node XV, represented by 16 clades. Donoghue and Moore (2003) recently asserted that all current methods of historical biogeographic analysis were susceptible to pseudo-congruence, but neither secondary BPA nor PACT suffer from that flaw. The other vicariant nodes include Node III, represented by 19 clades, corresponding to a vicariant split between the Sierra Madre del Sur + Transmexican Volcanic Belt and the areas to the north (Arizona, the Sonoran Desert, the Sierra Madre Occidental, the Southern Sierra Madre Occidental, and the Sierra Madre Oriental); Node X, represented by 14 clades, corresponding to a split between the Transmexican Volcanic Belt and the Sierra Madre del Sur; and Node VII, represented by 7 clades, corresponding to a split between Arizona + Sonoran Desert and the Sierra Madre Occidental + Southern Sierra Madre Occidental + Sierra Madre Oriental. A map of these vicariance events is shown in Fig. 13.
The remaining nine nodes depicted in Fig. 12 are biotic expansion nodes, corresponding to three distinct classes of dispersal episodes. The first of these comprises dispersal out of the Sierra Madre del Sur (area 7) primarily northward. The oldest of these is Node II, including 20 clades, and exhibits general dispersal, including some southward movement. More recent dispersal episodes out of the Sierra Madre del Sur include Node IX, including 15 clades, with dispersal primarily into the Transmexican Volcanic Belt and the Sierra Madre Occidental; and Node XI, including 9 clades, exhibiting general dispersal, primarily northward. The second class of dispersal episodes involves three sequential dispersal events out of the Sierra Madre Occidental + Sierra Madre Oriental: Node IV, including 18 clades, represents dispersal primarily into Arizona + North
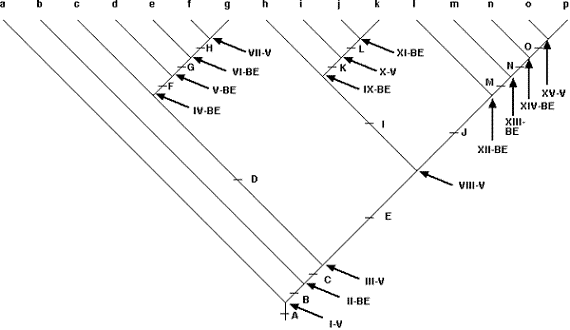
Figure 12. Partial representation of the area cladogram produced by secondary BPA of nine areas of endemism in Mexico and Central America, based on 33 clades used by Marshall & Liebherr (2000). Roman numerals denote the 15 general nodes, each supported by at least 7 clades, accompanied by an indication of whether the node indicates an episode of Vicariance (V) or Biotic Expansion (BE). Upper-case and lower-case letters refer to sub-area cladograms depicting the entire pattern of historical biogeographic diversity indicated by the members of the 33 clades. For details, conPACT corresponding author.

Figure 13. Map with vicariance splits plotted on it. AZ = Arizona; SD = Sonoran Desert; OCC = Sierra Madre Occidental; SOC = Southern Sierra Madre Occidental; ORI = Sierra Madre Oriental; TRAN = Sierra Transvolcanica; SUR = Sierra Madre del Sur; CGH = Chiapan Guatemalan Highlands; TAL = Talamancan Cordillera. (From Halas et al., 2005).
America, but with some southward dispersal; Node V, including 9 clades, represents dispersal in all directions and into all 9 areas, with successively fewer clades proceeding southward; and Node VI, including 8 clades, represents dispersal into Arizona + Sonoran Desert. Finally, there are three sequential dispersal events out of the Chiapan Guatemalan Highlands: Node XII, including 17 clades, represents dispersal primarily southward into the Talamancan Cordillera, but with some northward dispersal; Node XIII, including 19 clades, represents dispersal northward; and Node XIV, including 19 clades, represents dispersal northward, primarily into the Sierra Madre del Sur but with a few clades dispersing farther northward. The dispersal routes out of these three areas are depicted in Figures 14, 15, 16.
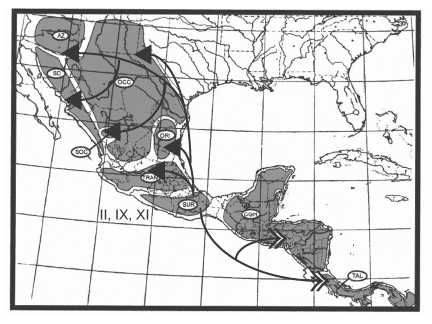
Figure 14. Dispersal from the Sierra Madre del Sur. Figures 14-16. Maps depicting dispersal routes out of three areas identified as sources of biotic expansion depicted in Fig.12. (From Halas et al., 2005).

Figure 15. Dispersal from the Sierra Madre Occidental + Sierra Madre Oriental. (From Halas et al., 2005).
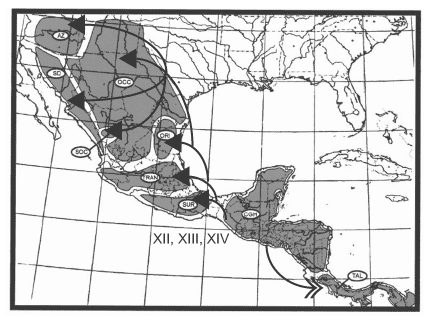
Oriental. 16. Dispersal from the Chiapan Guatemalan Highlands. Arrows with closed heads indicate primary dispersal routes; arrows with open heads indicate secondary dispersal routes. AZ = Arizona; SD = Sonoran Desert; OCC = Sierra Madre Occidental; SOC = Southern Sierra Madre Occidental; ORI = Sierra Madre Oriental; TRAN = Sierra Transvolcanica; SUR = Sierra Madre del Sur; CGH = Chiapan Guatemalan Highlands; TAL = Talamancan Cordillera.
Overall, nine of the 15 general nodes (60%) are biotic expansion nodes, involving general dispersal events from three areas (Sierra Madre del Sur, Sierra Madre Occidental + Sierra Madre Oriental, and Chiapan Guatemalan Highlands), and six (40%) are vicariance nodes, created by splits between the Transmexican Volcanic Belt + Sierra Madre del Sur and the Chiapan Guatemalan Highlands + Talamancan Cordillera (three times), between the Transmexican Volcanic Belt and the Sierra Madre del Sur (once), between the Transmexican Volcanic Belt + Sierra Madre del Sur and Arizona + Sonoran Desert + Sierra Madre Occidental + Southern Sierra Madre Occidental + Sierra Madre Oriental (once), and between Arizona + Sonoran Desert and the Sierra Madre Occidental + Southern Sierra Madre Occidental + Sierra Madre Oriental (once). In addition, with only a single exception (between nodes III and VIII), each vicariance node is separated by at least one biotic expansion node. This complex pattern of area relationships strongly supports an interpretation of taxon pulse-driven diversification for these biotas, with post-vicariance biotic dispersal producing widespread species that set the stage for succeeding episodes of vicariance.
Phylogenetic Inference of Modes of Initiating Speciation.
Assessing the modes of speciation for the members of the clades being analyzed is an important test of any hypothesis of taxon pulse-driven evolutionary radiation. If the general nodes interpreted as episodes of biotic expansion are evidence of taxon-pulse diversification, the more detailed portions of the area cladogram (denoted by upper- case and lower-case letters in Fig. 12), should exhibit clade-specific examples of (1) species with restricted ranges, (2) widespread species, and (3) clades of species produced by within-area speciation and by sequential peripheral isolates speciation.
When general nodes due to biotic expansion are differentiated from vicariant nodes, inferring speciation modes becomes complex. Vicariant speciation events are all those which occur at vicariance nodes (Fig. 12). Thus, all vicariant speciation in this data set is accounted for by the six nodes discussed in part II, along with an additional minor vicariance node in subtree a, splitting the Chiapan Guatemalan Highlands from the Talamancan Cordillera. There are three patterns associated with vicariance nodes: the clade undergoes a vicariant split (Fig. 17), the clade does not respond to the vicariance event (Fig. 18), so that there is only one species subsequent to the vicariance event, found in all vicariant areas, or the clade undergoes a split, followed by extinction in one of the vicariant areas (Fig. 19).

Figures 17-25. Historical biogeographic patterns used in inferring speciation modes. 6-11. Basal node indicates an inferred vicariance event splitting areas 1 and 2. 17. Vicariance only - species A and B are sister species occurring in areas 1 and 2, respectively. 18. Non-response to vicariance - species A and C are sister species occurring in areas 1 and 2, respectively, whereas species B is a member of a second clade, found in both areas 1 and 2. 19. Extinction - species A and B are sister species occurring in areas 1 and 2, respectively, whereas species C is a member of a second clade found only in area 2. The absence of a sistser species of C in area 1 is inferred to be due to extinction because the basal node is a vicariant node. 20. Vicariance splitting areas 1 and 2, with post speciation dispersal into area 1, followed by a new vicariance episode, again splitting areas 1 and 2, producing reticulated area relationships. Clade (A(BC)) shows both vicariant events; clade (DE) is inferred to have experienced both vicariance events and an extinction event in area 2; and clade (FG) is inferred not to have dispersed back into area 1, thus not experiencing the second vicariance event. 21. Vicariance + peripheral isolates speciation - species A and B are the result of vicariance splitting areas 1 and 2, with species C the result of dispersal into area 3. 22. Vicariance + within-area speciation - species A and B are the result of vicariance splitting areas 1 and 2, with species C the result of speciation within area 2. 23. Within-area speciation - the event producing species A and B occurred within areas 1+2. 24. Within-area speciation and post-speciation dispersal - - the event producing species A and B occurred within areas 1+2, with species A dispersing subsequently into area 4 and species B dispersing subsequently into area 6. 25. Peripheral isolates speciation producing reticulated area relationships - because the basal node is a biotic expansion node from area 1, species B is inferred to have been produced by dispersal from area 1 to area 2, and species C produced by dispersal from area 2 into area 1.
Asynchronous vicariance events splitting the same areas introduce an additional complication. Consider the following scenario: (a) a vicariance event splits two areas, 1 and 2, producing a pair of sister species, one in each area, (b) the species in area 2 subsequently disperses back into area 1, where (c) a second vicariant event between areas 1 and 2 occurs. Absence of a member of the clade in area 2 after the second vicariance event is still most parsimoniously counted as an inferred extinction event, but absence of a member of the clade in area 1 after the second vicariance event is more parsimoniously explained as a failure by that clade to disperse back into area 1 after the first vicariance event, in which case no inference of extinction is needed (Fig. 20).
Most of the vicariance nodes in the data set are followed by peripheral isolates or within-area speciation events. In counting species formed due to vicariance events, we do not infer that subsequent peripheral isolates or within-area speciation events cause the extinction of the vicariant species (Figs. 21, 22). In the case of a vicariant speciation event followed by a within-area speciation event (Fig. 22), it is impossible to determine, given our data, which of the descendant species represents the persistent ancestor; in such cases, we have assigned vicariance to one of the species arbitrarily for purposes of counting events. Actual determination of which, if any, of the two species in such cases represents the persistent ancestor would require more information about habitat heterogeneity, details of geographic distribution within the area, and information about ecological and/or behavioral divergence between sister species (Brooks and McLennan, 2002).
Biotic expansion nodes may include both peripheral isolates and within-area speciation events. At a biotic expansion node, if the range of a descendant species is outside that of the inferred ancestor at the node, the speciation event is counted as peripheral isolates. If the range of the descendant species includes that of the inferred ancestor at the node, the speciation is considered to be a within-area speciation
(Fig. 23). Throughout the data set, the existence of ancestor and descendant species in at least partially overlapping ranges is held to be evidence of within-area speciation; any discrepancy in ranges is counted as post-speciation dispersal (Fig. 24). This is because within-area speciation followed by dispersal is more parsimonious than peripheral isolates speciation followed by dispersal back into the ancestral range. An exception can occur at biotic expansion nodes, however. Since a biotic expansion is a coordinated event, in which multiple clades react to the same breakdown of a barrier, the ancestral range is determined for the tree as a whole, not for each individual clade. An individual clade may thus show a pattern which suggests within-area speciation but, upon comparison with the general expansion pattern, is better explained as peripheral isolates speciation followed by dispersal back into the ancestral range (Fig. 25). Note the similarity in pattern between figure 20 and figure 25, underscoring the importance of distinguishing vicariance nodes from biotic expansion nodes.
Cases in which a clade does not participate in a biotic expansion node are explained as a failure to disperse. While a vicariance event will necessarily split the species present in that area into two separate populations, allowing for speciation, the breakdown of a barrier at a biotic expansion node only creates the conditions that allow for dispersal. Extinction no doubt occurs among the clades which take part in biotic expansions, but it cannot be inferred based on the expansion pattern alone: further evidence is required to determine that an extinction event has taken place.
Biotic expansion nodes include species which have speciated sympatrically and then dispersed outward as the barrier associated with the expansion node broke down, forming a widespread species; species which dispersed following the breakdown of the barrier and speciated in the newly-colonized area, forming peripheral isolates species; and also species which dispersed outward, speciated, and then dispersed back into their ancestral range during a subsequent pulse, producing additional widespread species or peripheral isolates species. At minor nodes in sub-trees and along internodes, peripheral isolates and within-area speciation are distinguished in the same manner as at expansion nodes: if the range of a species overlaps at all with that of its inferred ancestor, it is counted as a within-area species; otherwise, it is considered a peripheral isolates species. Peripheral isolates speciation followed by within-area speciation creates the same problem as vicariance followed by peripheral isolates speciation. One of the two species following a within-area speciation must be counted as a persistent ancestor and the other as a species formed by peripheral isolates speciation. It is impossible to tell, without further data, which of the two species is the persistent ancestor, so the designation is made arbitrarily for the purposes of counting.
Within-area speciation is not restricted to sympatric speciation. Under the protocol used for determining mode of speciation, within-area speciation is almost always inferred when adjacent species in a clade co-occur in at least one area. Many such cases, however, are likely to be episodes of vicariance and peripheral isolates speciation occurring on spatial scales smaller than those of the areas used by Marshall and Liebherr (2000). Another possibility for widespread species explained as within-area speciation is peripheral isolates speciation followed by postspeciation dispersal back into the ancestral area. For each case in which sister species partially overlap in range, it is more parsimonious to assume that within-area speciation occurred in the shared area, followed by post-speciation dispersal, than it is to assume that an initial episode of dispersal occurred, producing peripheral isolates speciation, followed by a second episode of dispersal, back into the ancestral range. It is unrealistic to think, however, that cases of peripheral isolates speciation with subsequent dispersal never occur. Indeed, the taxon pulse model gives additional reason to assume that such events do, in fact, occur. Under the taxon pulse model, species undergo periods of dispersal when barriers break down, followed by contraction and speciation as barriers reform; dispersal occurs again as the new barriers break down. A population could thus disperse to a new area during the initial expansion phase, become isolated and speciate during the contraction phase, and then disperse back into the range of its ancestor during the second expansion phase. Recognizing such cases requires additional information about geographic distributions within each area, likely resulting in further sub-division of the areas of endemism used by Marshall and Liebherr (2000), as well as information about habitat heterogeneity within each area and episodes of ecological diversification associated with particular speciation events (for protocols, see Brooks and McLennan, 2002).
A total of 56% of the nodes in the taxon-area cladograms in the Marshall and Liebherr (2000) data pertain to within-area speciation events; hence, inferences of among-area relationships are based on only 44% of the speciation events in the data set. That 44% is further divided between vicariance (19%) and peripheral isolates speciation (25%). The inferences about speciation, therefore, also support the predictions of taxon pulse-driven diversification rather than vicariance-driven diversification.
Phylogenetic Influences on Species-Area Relationships in the Assemblage of Biotas.
That larger islands have greater species richness than smaller islands, and that «islands» need not be oceanic because species richness increases with any increased sample of area, has long been recognized. This increase follows a simple power function mathematically expressed as S=cAz (Preston, 1962). Figure 26 shows the results of correlating species richness and area size for the 33 clades and 9 areas in Mexico and Central America examined herein (for details see Halas et al., 2005). The low correlation coefficient for the species-area curve (r2 =0.47) is due primarily to relatively small areas containing unusually large numbers of species.
The Equilibrium Theory of Island Biogeography (ETIB: MacArthur and Wilson, 1963, 1967) predicts a linear relationship between species richness and the size of an island resulting from a dynamic balance between immigration, that is, colonization from a source area, and extinction. The extinction rate is assumed to increase with the number of species present on any island, so that small areas with higher species richness than expected have a higher extinction rate than immigration rate and are not yet in equilibrium. Correlating extinction events and species richness for this data set (Fig. 27) produces a high correlation coefficient (r2 =0.75), indicating strong support for this prediction of the ETIB.
Immigration rate to any given island is expected to decrease with increasing species richness, reaching zero when all species from the source area have colonized the island. As immigration is the only source of new species, we would therefore expect species richness to increase with number of colonization events. Correlating colonization events (peripheral isolates speciation + post-speciation dispersal) and species richness for this data set (Fig. 28), however, produces a relatively low correlation coefficient (r2 = 0.36). This suggests that colonization is not the primary mechanism contributing to species richness in these areas. This is underscored when colonization events are correlated with area size (Fig. 29), which produces a very low correlation coefficient (r2 = 0.05), separately from in situ speciation events (vicariance + within-area speciation) and area size (Fig. 30), which produces a much higher correlation coefficient (r2 =0.60). It is clear that in situ speciation contributes more to the species-area relationship than does colonization.
The original mathematical expression of the ETIB is: change in species number, «s, equals immigration, M, plus within-area speciation, G, minus species extinction, D, or «s = M + G - D. MacArthur and Wilson (1963: 380) stated, however, that «for most cases it was probably safe to omit G from the model» as the effect of in situ speciation on the species-area relation is «probably significant only in the oldest, largest, and most isolated islands.» MacArthur and Wilson (1963) acknowledged that ‘local speciation‘ (in situ speciation) would confound the species-area relationship, and more recent discussions have suggested that such historical phenomena require closer investigation (e.g., Heaney, 2000; Whittaker, 2000). Losos and Schluter (2000) reported that, for Anolis lizards on large Caribbean islands, inferred extinction rates were low and in situ speciation was a more important source of species richness than colonization. They predicted that effects similar to those they observed on the largest islands should be found on continental islands. The results for 33 clades on large continental islands corroborate all those predictions: in situ speciation correlates better with area size than does colonization, and inferred extinction rates are low. The protocol produces inferences of 19 extinction events involving 16 of the 33 clades. This is a minimal inference of extinction rate, because it permits parsimonious inferences of extinction associated only with episodes of vicariance. If extinction rates are the same following biotic expansion events, which account for 60% of the general nodes, the number of inferred extinctions increases only from 19 to 48, compared with 333 observed species inferred to be the product of 281 speciation events.
These data provide additional insight into the evolutionary relationship between colonization and in situ speciation. There is a very poor correlation between colonization and in situ speciation (Fig. 31; r2 = 0.02), indicating that these phenomena are relatively independent of each other. We suggest that the reason historical effects on large islands confound the species-area relationship is not colonization or the in situ production of species per se, but rather the subsequent dispersal of some species produced in situ to other islands, so that sources become islands and islands become sources on evolutionary time scales. All nine areas discussed above have been colonized and have produced colonizers, thus acting as both sources and islands at different times and to different degrees. Even the areas identified in figure 12 as sources of biotic expansion events, the Sierra Madre del Sur, the Sierra Madre Occidental + Sierra Madre Oriental, and the Chiapan Guatemalan Highlands, have acted as islands for colonization. The Transmexican Volcanic Belt has acted as an «island», receiving species by colonization, for all 9 biotic expansion nodes in figure 12, whereas the Sierra Madre del Sur has acted as a dispersal source for three of the biotic expansion nodes in figure 12. At the same time, both areas were involved in five of the six vicariance nodes in figure 12. This explains why these relatively small areas are disproportionately species-rich, without any evidence of an accompanying high extinction rate.
Conclusions
The maximum vicariance hypothesis, and all methods of historical biogeographic analysis stemming from it, have produced an inadequate and inaccurate representation of historical biogeographic patterns and processes. This is due to the overly simplistic nature of the underlying model of maximum vicariance, and the overly restrictive range of processes permitted by the methods designed to represent historical biogeographic patterns. Although the model of maximum vicariance has been falsified, vicariance remains an integral part of historical biogeography. The taxon pulse hypothesis proposes that biotic evolution is the result of alternating episodes of vicariance and biotic expansion, each producing general patterns of geographic relationships. Recent empirical studies, using secondary BPA and PACT, which provide an accurate depiction of biogeographical patterns and which are less restrictive in terms of permitted processes, corroborate the taxon pulse hypothesis. I therefore propose that the taxon pulse be considered the new general model for historical biogeography.
In addition to providing a more accurate representation of historical patterns, PACT permits historical biogeography, represented by the taxon pulse, and ecological biogeography, represented by the ETIB, to begin the long-overdue process of integration, which almost occurred in the mid-1980s (Brooks, 2004). This complements recent calls by, e.g., Heaney (2000), and Whittaker (2000) for modifications of the ETIB to incorporate complex patterns of immigration, extinction, and diversification occurring on various spatial scales and on both ecological and evolutionary time scales.
Acknowledgments
I express special thanks to the Organizing Comitee of the «Primera Reunión Mexicana de Biología Filogenética» Xalapa, Mexico, June, 2004, particularly Ana Barahona and Gerardo Pérez Ponce de León for inviting me to present these ideas. I also owe a special debt of gratitude to Deborah McLennan (University of Toronto), Rick Winterbottom (Royal Ontario Museum), Eric Hoberg (US National Parasite Collection), Rino Zandee (Leiden University), Marco Van Veller (Wageningen University), Bruce Lieberman (University of Kansas), Brian Crother (Southeastern Louisiana University), Brett Riddle (University of Nevada-Las Vegas), Ashley Dowling (University of Michigan), and Maggie Wojcicki, David Zamparo, Dominik Halas, Mike Spironello, and Marc Green (University of Toronto) for helpful discussions and critical insights. This study was supported by a grant from the Natural Sciences and Engineering Research Council (NSERC) of Canada.
Literature cited
Bouchard, P., D. R. Brooks, and D. K. Yeates. 2004. Mosaic macroevolution in Australian wet tropics arthropods: Community assemblage by taxon pulses. In Rainforest: Past, Present, Future, C. Moritz and E. Bermingham (eds.), University of Chicago Press, Chicago. pp 425-469. [ Links ]
Brooks, D. R. 1981. Hennig's Parasitological method: A proposed solution. Systematic Zoology 30: 229-249. [ Links ]
Brooks, D. R. 1990. Parsimony analysis in historical biogeography and coevolution: methodological and theo- retical update. Systematic Zoology 39: 14-30. [ Links ]
Brooks, D. R. 2004. Reticulations in historical biogeogra- phy: the triumph of time over space in evolution. In Frontiers of Biogeography. New Directions in the Geo- graphy of Nature, M. V. Lomolino and L. R. Heaney (eds.). Sinauer Associates, Sunderland,Massachusetts. pp. 125-144. [ Links ]
Brooks, D. R. and McLennan, D. A. 1991. Phylogeny, Eco- logy and Behavior: A Research Program in Comparative Biology. University of Chicago Press, Chicago. 434 p. [ Links ]
Brooks, D. R. and McLennan, D. A. 2001. A comparison of a discovery-based and an event-based method of historical biogeography. Journal of Biogeography 28:757-767. [ Links ]
Brooks, D. R. and McLennan, D. A. 2002. The Nature of Diversity: An Evolutionary Voyage of Discovery. Univer- sity of Chicago Press, Chicago,676p. [ Links ]
Brooks, D. R. and McLennan, D. A. 2003. Extending phy- logenetic studies of coevolution: secondary Brooks par- simony analysis, parasites, and the Great Apes. Cladistics 19: 104-119. [ Links ]
Brooks, D. R., Van Veller, M. G. P., and McLennan, D. A. 2001. How to do BPA, Really. Journal of Biogeography 28: 345-358. [ Links ]
Brundin, L. 1966. Transantarctic relationships and their significance, as evidenced by chironomid midges. Kungliga Svenska Vetenskaps Akademiens Handlingar, FjSrde Serien11: 1-472. [ Links ]
Brundin, L. 1972. Evolution, causal biology and classifica- tion. Zoologica Scripta 1: 107-120. [ Links ]
Cormen, T. H., C. E. Leiserson,R. L. Rivest, and S. Clifford. 2001. Introduction to algorithms. McGraw-Hill, Toronto, 1184 p. [ Links ]
Cressey, R. F., B. Collette, and J. Russo. 1983. Copepods and scombrid fishes: A study in host-parasite relationships. Fisheries Bulletin 81: 227-265. [ Links ]
Darlington, P. J. 1943. Carabidae of mountains and islands: data on the evolution of isolated faunas, and on atrophy of wings. Ecological Monographs 13: 37-61. [ Links ]
Darwin, C. 1872. Origin of Species. 6 th ed., John Murray, London,502 p. [ Links ]
Donoghue, M. J. and Moore, B. R. 2003. Toward an integra- tive historical biogeography. Integrative and Comparative Biology 43: 261-270. [ Links ]
Dowling, A. P. G. 2002. Testing the accuracy of TreeMap and Brooks parsimony analyses of coevolutionary patterns using artificial associations. Cladistics 18: 416-435. [ Links ]
Dowling, A. P. G., M. G. P. van Veller, E. P. Hoberg, and D.R. Brooks. 2003. A priori and a posteriori methods in comparative evolutionary studies of host-parasite asso- ciations. Cladistics 19: 240-253. [ Links ]
Endler, J. A. 1982. Problems in distinguishing historical from ecological factors in biogeography. American Zoo-logist 22: 441-452. [ Links ]
Erwin, T. L. 1979. Thoughts on the evolutionary history of ground beetles: hypotheses generated from comparative faunal analyses of lowland forest sites in temperate and tropical regions. In Carabid Beetles: Their Evolution, Natural History, and Classification, T. L. Erwin, G. E. Ball and D. R. Whitehead (eds.), W. Junk Eds., The Hague, pp. 539-592. [ Links ]
Erwin, T. L. 1981. Taxon pulses, vicariance, and dispersal: an evolutionary synthesis illustrated by carabid beetles. In Vicariance Biogeography - A Critique, G. Nelson and D. E. Rosen, (eds.), Columbia University Press, New York, pp. 159-196. [ Links ]
Green, M., M. G. P. Van Veller, and D. R. Brooks. 2002. Assessing modes of speciation: range asymmetry and biogeographical congruence. Cladistics 18: 112-124. [ Links ]
Halas, D., D. Zamparo, and D. R. Brooks. 2005. A protocol for studying biotic diversification by taxon pulses. Journal of Biogeography 32: 249-260. [ Links ]
Heaney, L. R. 2000. Dynamic disequilibrium: a long-term, large-scale perspective on the equilibrium model of island biogeography. Global Ecology and Biogeography 9: 59- 74. [ Links ]
Humphries C. J. and L. R. Parenti. 1999. Cladistic Biogeo- graphy. Second edition: Interpreting patterns of plant and animal distributions. Oxford University Press, New York. 187 p. [ Links ]
Lieberman, B. S. 2000. Paleobiogeography. Plenum/Kluwer Academic, New York,222 p. [ Links ]
Lieberman, B. S. 2003a Unifying theory and methodology in biogeography. Evolutionary Biology 33: 1-25. [ Links ]
Lieberman, B.S. 2003b. Paleobiogeography: The relevance of fossils to biogeography. Annual Review of Ecology,Evolution, and Systematics 34: 51-69. [ Links ]
Losos, J. B. and Schluter, D. 2000. Analysis of an evolu- tionary species-area relationship. Nature 408: 847- 850. [ Links ]
MacArthur, R. H. and Wilson, E. O. 1963. An equilibrium theory of insular zoogeography. Evolution 17: 373-387. [ Links ]
MacArthur, R. H. and Wilson, E. O. 1967. The Theory of Island Biogeography. Princeton University Press Princeton, N. J. 203 p. [ Links ]
Marshall, C. J. and Liebherr, J. K. 2000. Cladistic biogeo- graphy of the Mexican transition zone. Journal of Biogeo- graphy 27: 203-216. [ Links ]
McLennan, D. A. and Brooks, D. R. 2002. Complex histories of speciation and dispersal: An example using some Australian birds. Journal of Biogeography 29:1055-1066. [ Links ]
Nelson, G. and Platnick, N. I. 1981. Systematics and Bio- geography: Cladistics and Vicariance. Columbia University Press, New York,567p. [ Links ]
Nelson, G. and Rosen, D. E. (eds). 1981. Vicariance Bio- geography: A Critique. Columbia University Press, New York,593 p. [ Links ]
Page, R. D. M. 1990. Temporal congruence and cladistic analysis of biogeography and cospeciation. Systematic Zoology 39: 205-226. [ Links ]
Platnick, N. I. and Nelson, G. 1978. A method of analysis for historical biogeography. Systematic Zoology 27: 1- 16. [ Links ]
Porzecanski, A.L. and J. Cracraft. 2005. Cladistic analysis of distributions and endemism (CADE): using raw distributions of birds to unravel the biogeography of the South American aridlands. Journal of Biogeography 32: 261- 275. [ Links ]
Preston,F. W. 1962. The canonical distribution of common- ness and rarity: Part I. Ecology 43: 185-215. [ Links ]
Ronquist, F. 2002. Parsimony analysis of coevolving species associations. In Tangled Trees R. D. M. Page (ed.), University of Chicago Press, Chicago, pp 22-64. [ Links ]
Siddall, M. E. and Perkins, S. L. 2003. Brooks Parsimony Analysis: A valiant failure. Cladistics 19: 554-564. [ Links ]
Simberloff, D. 1987. Calculating probabilities that clado- grams match: a method of biogeographical inference.Systematic Zoology 36: 175-195. [ Links ]
Simberloff, D., K. L. Heck, E. D. McCoy, and E. F. Connor. 1981. There have been no statistical tests of cladistic biogeographic hypotheses. In Vicariance Biogeography: A Critique, G. Nelson and D. E. Rosen (eds.): Columbia University Press, New York pp. 40-63. [ Links ]
Simpson, G. G. 1944. Tempo and Mode in Evolution. New York: Columbia University Press. 242 p. [ Links ]
Soest, R. W. M. van and E. Hajdu. 1997. Marine area rela- tionships from twenty sponge phylogenies. A comparison of methods and coding strategies. Cladistics 13: 1- 20. [ Links ]
Spironello, M. and D. R. Brooks. 2003. Dispersal and diversification in the evolution of Inseliellium,an archi- pelagic dipteran group. Journal of Biogeography 30: 1563- 1573. [ Links ]
Van Veller, M. G. P. and D. R. Brooks. 2001. When simpli- city is not parsimonious: A priori and a posteriori methods in historical biogeography. Journal of Biogeography 28: 1-11. [ Links ]
Van Veller, M. G. P., D. R. Brooks, and M. Zandee. 2003. Cladistic and phylogenetic biogeography: the art and the science of discovery. Journal of Biogeography 30: 319 329. [ Links ]
Van Veller, M. G. P., D. J. Kornet, and M. Zandee. 2000. Methods in vicariance biogeography: assessment of the implementation of assumptions zero, 1 and 2. Cladistics 16: 319-345. [ Links ]
Van Veller, M. G. P., D. J. Kornet, and M. Zandee. 2001 A posteriori and a priori methodologies for testing hypotheses of causal processes in vicariance biogeography. Cladistics 17: 248-259. [ Links ]
Van Veller, M. G. P., M. Zandee, and D. J. Kornet. 1999. Two requirements for obtaining valid common patterns under different assumptions in vicariance biogeography. Cladistics 15: 393-406. [ Links ]
Van Veller, M. G. P., M. Zandee, and D. J. Kornet. 2002. Testing hypotheses regarding general patterns in vica- riance biogeography with a posteriori and a priori methods. Cladistics 18: 207-217. [ Links ]
Whittaker, R. 2000. Scale, succession and complexity in island biogeography: are we asking the right questions? Global Ecology and Biogeography 9: 75-85. [ Links ]
Wiley, E. O. 1981. Phylogenetics: The Theory and Practice of Phylogenetic Systematics. Wiley and Sons, New York, 439 p. [ Links ]
Wiley, E. O. 1986. Methods in vicariance biogeography. In Systematics and Evolution P. Hovenkamp (ed.). University of Utrecht Press, Utrecht,pp. 283-306. [ Links ]
Wiley, E. O. 1988a. Parsimony analysis and vicariance biogeography. Systematic Zoology 37: 271-290. [ Links ]
Wiley, E. O. 1988b. Vicariance biogeography. Annual Review of Ecology and Systematics 19: 513-542. [ Links ]
Wilson, E. O. 1959. Adaptive shift and dispersal in a tropical and fauna. Evolution 13: 122-144. [ Links ]
Wilson, E. O. 1961. The nature of the taxon cycle in the Melanesian ant fauna. American Naturalist 95: 169-193. [ Links ]
Wojicki, M. and D. R. Brooks. 2005. PACT: A simple and efficient algorithm for generating area cladograms. Journal of Biogeography. 32: 755- 774. [ Links ]
Zandee, M. and M. C. Roos. 1987. Component-compatibility in historical biogeography. Cladistics 3: 305-332. [ Links ]














