Introduction
Substrate specificity is instrumental in enzyme catalysis and a major determinant of the enzyme’s physiological role. Given these facts, much effort has been devoted to understanding the factors that define which compound or compounds can be used as substrates by a determined enzyme. Enzymes may be strictly specific for one substrate or exhibit a certain degree of substrate promiscuity, the latter being used by evolution for the emergence of new enzymes that perform novel metabolic roles [1]. Here, we exemplify that specificity may exist within promiscuity taking as an example the four aminoaldehyde dehydrogenases from Pseudomonas aeruginosa.
The aldehyde dehydrogenase superfamily comprises enzymes that catalyze the NAD(P)+-dependent oxidation of a great variety of endogenous or exogenous aldehydes. Thus, they participate in important catabolic and anabolic pathways and in detoxification processes [2]. ALDHs are considered to exhibit a rather broad specificity towards their aldehyde substrates, although some of them are highly specific. The aminoaldehyde dehydrogenases (AMADHs) form a particular ALDH subgroup that, as their name indicates, oxidize aldehydes bearing a primary, secondary, tertiary or quaternary amino group. In vitro, many AMADHs can use most of the aminoaldehydes as substrates, but, despite this apparent promiscuity, their selectivity for their substrates exquisitely suits their in vivo roles. In accordance with its known metabolic versatility, P. aeruginosa possesses four AMADHs: PA5373-also called PaBADH-, PA5312-named PaPauC and formerly PaKauB-, PA4189, and PA0219, which participate in different metabolic pathways (as a comparison, humans only have one). PA5373 belongs to the ALDH9 family [3] and PA5312, PA4189 and PA0219 belong to the ALDH27 family, which is exclusive of bacteria [4,5]. Hence, the degree of identity between the three ALDH27 enzymes is greater than that of the ALDH9 enzyme (Fig. 1). It is known that PA5373 oxidizes betaine aldehyde (BAL) [6] and is involved in choline degradation and in the synthesis of the osmoprotectant glycine betaine [7,8]. PA5312 is the ALDH oxidizing 3-aminopropionaldehyde (APAL), 4-aminobutyraldehyde (ABAL), and their derivatives glutamyl-3-aminopropionaldehyde (GluAPAL) and glutamyl-4-aminobutyraldehyde (GluABAL), all of them formed in the catabolism of polyamines [9-11]. It also oxidizes 4-guanidinobutyraldehyde (GdBAL) formed in the degradation of arginine [12]. Phenotypic, transcriptional, and metabolomic studies suggested that PA0219 oxidizes 4-imidazole acetaldehyde (ImCTAL) in a hypothetical histamine degradative pathway [13,14]. The physiological role of PA4189 remains unknown, although this enzyme has been recently biochemically and structurally characterized in our group [15] and found that it efficiently oxidizes aminoacetaldehyde (ACTAL) formed in a so far uncharacterized metabolic pathway. The reaction of oxidation of these aminoaldehydes possible substrates of the P. aeruginosa AMADHs is schematically depicted in Fig. 2.
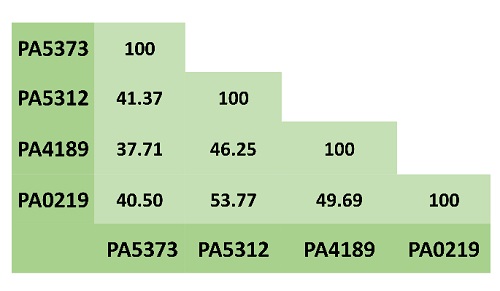
Fig. 1 Amino acid sequence identity between the four P. aeruginosa AMADHs. Multiple alignments were performed with BLASTP (https://blast.ncbi.nlm.nih.gov/).
Experimental
Chemicals and biochemicals
HEPES, EDTA, BAL chloride, the diethylacetal forms of ACTAL, APAL and ABAL, 2-mercaptoethanol, NAD+, and NADH were obtained from Sigma-Aldrich. GluAPAL and GluABAL diethylacetals were synthesized as previously reported [11], and GdBAL as described in [16]. The exact concentrations of NADH and NAD+ were measured by their respective absorbance at 340 nm ( 340 = 6,200 M-1 cm-1) and 260 nm ( 260 = 18,000 M-1 cm-1). ACTAL, APAL, ABAL, GluAPAL, and GluABAL were prepared freshly, hydrolyzing the corresponding diethylacetals as described. [11,15]. The exact concentration of the aldehyde was measured in each experiment by determining the amount of NADH produced after its complete oxidation by using the corresponding pure AMADH enzymes and the standard assay conditions described below.
Enzyme purification and activity assays
PA5373 was purified from P. aeruginosa cells as reported [6]. Recombinant PA5312 and PA4189 were over-expressed as His-tagged proteins and purified to homogeneity as previously described [11,15]. PA0219 was produced as a His-tagged recombinant protein in BL21RIL E. coli cells (Agilent) transformed with the plasmid pET28b:PA0219, which contains the full sequence of the PA0219 gene, and purified following the same procedure described for PA4189. The enzymes purity was assessed by SDS-PAGE according to Laemmli [17]. Proteins concentration was determined spectrophotometrically, using their molar absorptivity at 280 nm predicted from amino acid sequence with ExPASy ProtParam [18], which are 52,060 M−1 cm−1 for PA5373, 69,900 M-1 cm-1 for PA5312, 55,920 M−1 cm−1 for PA4189, and 63,940 M−1 cm−1 for PA0219.
The AMADHs activity was measured spectrophotometrically at 30 °C by monitoring the increase in the absorbance at 340 nm in a mixture (0.5 mL) consisting of 100 mM potassium phosphate buffer (or 150 mM pyrophosphate buffer in the case of PA4189), pH 8, 1 mM aldehyde, and 0.6 mM NAD+. All assays were initiated by addition of the enzyme. One unit of activity is defined as the amount of enzyme that catalyzes the formation of 1 (mol of NADH per min under our assay conditions.
Amino acid sequence and structural comparisons
Amino acid sequence alignments of the four P. aeruginosa AMADHs were performed with BLASTP (https://blast.ncbi.nlm.nih.gov/) and Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/). AMADHs amino acid sequences were obtained at the National Center of Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/protein/) with the following protein ID: NP254060.1 for PA5373, NP253999.1 for PA5312, NP252878.1 for PA4189, and NP248910.1 for PA0219. Structural comparisons were performed with ChimeraX [19], using the crystallographic structures in the RSCB Protein Data Base (PDB; https://www.rcsb.org/) with codes 2WME for PA5373, 6B4R for PA5312, and 7UYY for PA4189. For PA0219 we used the model obtained from the AlphaFold Protein Structure Database (https://alphafold.com/) [20] with code Q9I6R9.
Results and discussion
The best in vitro aldehyde substrates of the P. aeruginosa AMADHs
With comparative purposes, the specific activity of the four P. aeruginosa AMADAHs was determined at 1 mM of the aldehydes reported as, or suggested to be, their physiological substrates and 0.6 mM NAD+, which is a concentration considered close to the physiological one [21]. We did not assay ImCTAL-the proposed PA0219 physiological substrate- because our repeated efforts to synthesize it have failed so far.
As shown in Fig. 3 and Table 1 the reported physiological substrate or substrates for PA5373 (BAL) [6-8], PA5312 (GdBAL, GluAPAL, GluABAL, ABAL, and APAL) [11], and PA4189 (ACTAL) [15] were the best in vitro substrates for each of these enzymes. On the contrary, they were poor substrates for the other AMADHs, particularly BAL, clearly indicating the need for an enzyme highly specific for this aldehyde, which is fulfilled by PA5373. PA5312 is the P. aeruginosa AMADH with the broadest substrate specificity, consistent with its participation in several polyamine degradation routes (11 and references therein) and one pathway of arginine degradation [12]. PA4189 appears highly specific for ACTAL, although the metabolic pathway in which this enzyme participates-and in which ACTAL would form as an intermediate-has not been characterized yet. Under our assay conditions, PA0219 exhibited the highest activity with APAL, followed by GdBAL and ABAL. However, its activity with these aldehydes is lower than that of PA5312, particularly with GdBAL and ABAL, suggesting that none of them is the PA0219 physiological substrate. Additionally, other good substrates of PA5312 were very poor substrates of PA0219. Clearly, to obtain a complete picture of the aldehyde specificity of PA0219, it remains to test its activity with its putative physiological substrate, ImCTAL, and compare this activity with those of the others P. aeruginosa AMADHs.

Fig. 3 Aminoaldehyde specificity of the four P. aeruginosa AMADHs. Specific activity was determined at 1 mM of the different aminoaldehydes and 0.6 mM NAD+ at 30 oC and pH 8.0. Data are the mean ( S.D. values obtained using two enzyme purification batches and at least two technical replicates differing no more than 10 %. For clarity of the figure, the BAL data are shown separately. ACTAL: aminoacetaldehyde; APAL: 3-aminopropionaldehyde; ABAL: 4-aminobutyraldehyde. BAL: betaine aldehyde; GluAPAL: glutamyl-3-aminopropionaldehyde; GluABAL: glutamyl-4-aminobutyraldehyde; GdBAL: 4-guanidinobutyraldehyde.
Table 1 Activitya of P. aeruginosa AMADHs with aminoaldehydes.
| SPECIFIC ACTIVITY (U mg protein-1) | ||||
| ALDEHYDES | PA5373 | PA5312 | PA4189 | PA0219 |
| ACTAL | 0.02 ± 0.00 | 7.4 ± 0.2 | 40.6 ± 1.6c | 0.18 ± 0.02 |
| APAL | 3.8 ± 0.2 | 11.5 ± 0.3b | 6.5 ± 0.4c | 5.0 ± 0.0 |
| ABAL | 0.69 ± 0.02 | 20.9 ± 0.3b | 1.5 ± 0.0c | 2.4 ± 0.0 |
| BAL | 122 ± 4 | 0.05 ± 0.00 | 0.03 ± 0.00c | 0.02 ± 0.01 |
| GluAPAL | 2.2 ± 0.1 | 30.1 ± 2.5b | 3.3 ± 0.1c | 0.40 ± 0.01 |
| GluABAL | 2.8 ± 0.1 | 26.4 ± 0.3b | 0.08 ± 0.02c | 0.65 ± 0.00 |
| GdBAL | 0.56 ± 0.05 | 41.6 ± 0.8 | 0.03 ± 0.00c | 0.02 ± 0.01 |
aSpecific activities were determined at pH 8.0, 30 oC, 1 mM aldehyde and 0.6 mM NAD+. Data are the mean ( S.D. of initial velocity values obtained using two enzyme purification batches and at least two technical replicates differing no more than 10 %. bValues were taken from [11]. cValues were taken from [15].
The above-described results indicate that the four P. aeruginosa AMADHs have experienced selective pressures to acquire the active-site features that define their respective substrate specificity, not only by being highly active with some aldehydes, but also by being quite inactive with others. Bellow, we explore these structural features by comparatively analyzing the active sites of these four enzymes.
The active-site of the P. aeruginosa AMADHs
To structurally compare the active site of the four AMADHs, we used the crystallographic structures deposited by our group in the PDB with accession codes 2WME (PA5373), 6B4R (PA5312), and 7UYY (PA4189), and the AlphaFold model Q9I6R9, since the PA0219 crystal structure has not been determined yet. Although we also obtained the PA0219 tridimensional-structure model proposed by RoseTTAfold [22] (www.robetta.org) and found that was similar to the AlphaFold model-the (-carbons average root medium square deviation (r.m.s.d.) value between these two models is 0.779 Å-, we decided to use the AlphaFold model in our studies because of significant differences between the two models in the conformation of important active-site residues (Fig. 4). Thus, the Phe465 sidechain is away from the active site in the RoseTTAfold model, while in the AlphaFold model is directed toward the active site in a similar conformation to that observed in the PA5312 and PA4189 crystal structures. Also, in the RoseTTAfold model, the sidechain of the catalytic Asn166 is in a conformation not observed before in any ALDH crystal structure and incompatible with this residue role during the catalyzed reaction.

Fig. 4 Comparison of the sidechain conformation of active-site residues in the AlphaFold and RoseTTAfold models of PA0219. The figure was constructed with ChimeraX [19].
As in every ALDH, the P. aeruginosa AMADHs have an essential catalytic cysteine residue, whose thiol group performs the nucleophilic attack on the carbonyl carbon of the aldehyde substrates in the first step of their catalytic mechanism [23]. The sidechain of this residue was observed in the so- called “resting” conformation [24] in the crystal structures of PA5373 and PA5312, and in the so-called “attacking” conformation [24] in the crystal structure of PA4189 and in the RoseTTAfold and AlphaFold models (Fig. 4). The two others catalytic residues present in these enzymes are: 1) The conserved asparagine residue that stabilizes the thiohemiacetal oxyanion through a hydrogen bond with the amide nitrogen of its sidechain [25 and references therein]. In the three crystal structures and in the PA0219 AlphaFold model, this residue is in the conformation observed in the rest of the ALDH crystals reported to date. 2) The glutamate that activates the water molecule needed in the hydrolysis of the thioester to release the acid product of the reaction [25 and references therein] is only conserved in the hydrolytic ALDHs, such as the ones studied here. This residue was observed in the “intermediate” conformation in every known P. aeruginosa AMDH crystal and in the PA0219 AlphaFold model, although in PA5312 the “inside” conformation with occupancy of 0.46 and in PA5373 the “outside” conformation with 0.2 occupancy were observed (not shown).
Additionally, in the active site of every ALDH enzyme there are non-catalytic residues that play important roles in the recognition and specific binding of the aldehyde substrate. The nature of these residues depends of the chemical properties of the aldehyde for which these enzymes are specific. Given the basicity of their amino group, most aminoaldehydes bear a positive charge at the physiological pH of P. aeruginosa, which has been estimated to be 7.8 [26]. Therefore, their amino groups could make ionic bonds with the negatively charged carboxylic group of an acidic residue, glutamate or aspartate. Also, a particular arrangement of two or three aromatic residues, forming which has been called an “aromatic box” [27], has been reported to contribute to the binding of the positively charged amino group through cation-( interactions in AMADHs from other organisms [28]. Accordingly, in the active site of the four P. aeruginosa AMADHs, there are an acidic residue-aspartate in PA5312 and PA0219, glutamate in PA5373 and PA4189-and two aromatic residues with similar conformations, as shown in Fig. 5. There is a third aromatic residue in the PA5312, PA4189, and PA0219 active sites but not in PA5373, where the residue at the equivalent position is a valine. Interestingly, the latter aromatic residue-the phenylalanine at position 467, 464, and 465 in the amino acid sequences of PA5312, PA4189, and PA0219, respectively-and the active-site acidic residues belong to the so-called “anchor” loop [28], which is present in every ALDH and has been shown to comprise residues defining the aldehyde specificity of these enzymes [29-31].

Fig. 5 The acidic and aromatic residues in the active site of the P. aeruginosa AMADHs. Cartoon representation of the mainchain with active site residues as sticks. The mainchain carbons are colored in grey, except those of anchor loop, which are colored in green. Sidechains atoms are colored in red (the acid residues), turquoise (the aromatic residues), yellow (the catalytic cysteine), and purple (the Val453 of PA5373). The figure was constructed with ChimeraX [19], using the crystal structures with PDB accession codes 2WME (PA5373) (a), 6B4R (PA5312) (b), 7YUU (PA4189) (c), and the AlphaFold model Q9I6R9 (PA0219) (d).
The sidechains of the PA5312 and PA0219 aspartate residues and the PA4189 glutamate residue are inside their respective active sites, but that of the PA5373 glutamate is away from the active site (Fig. 5(a)), and therefore cannot participate in the binding of the aldehyde. Also, in PA5373 the residue equivalent to Asp459 and Asp457 in PA5312 and PA0219, respectively, is a glycine, and the anchor loop’s phenylalanine of the three ALDH27 enzymes is a valine, as mentioned above. Therefore, neither of these residues can interact with the aminoaldehyde substrate in PA5373. These three differences of the PA5373 active site play an important role in determining its high activity with BAL, because they allow the entering of this aldehyde deep into the active site, where its trimethylammonium group interacts with the other two aromatic residues-Tyr154 and Trp161, PA5373 numbering)-and its carbonyl group can adopt the proper position for catalysis. Our results of manual dockings of BAL in the active site of the four enzymes (Fig. 6) support this and suggest that the very low activity of the other three enzymes with this aldehyde (Fig. 3) is due to steric clashes of the BAL bulky trimethylammonium group with the sidechains of the acidic residue and an opposing leucine residue that narrow the aldehyde-entrance tunnel in PA5312 and PA4189, or the sidechains of a leucine(also belonging to the anchor loop(and an opposing isoleucine residue in PA0219, although in this enzyme there is some incertitude since the analyzed structure is not crystallographic. Because of these clashes, the BAL aldehyde group cannot reach the thiol group of the catalytic cysteine and, therefore, the nucleophilic attack cannot take place. In PA5373, as a result of the absence of a sidechain in the residue at equivalent to Asp459 and Asp457 in PA5312 and PA0219, respectively, and the position of the Glu446 sidechain away from the active site, there is no steric hindrance for the entering of BAL and its catalytic correct positioning inside the active site of this enzyme.

Fig. 6 Manual dockings of BAL into the active site of the four P. aeruginosa AMADHs showing clashes of BAL with active site residues in all of them except PA5373. BAL atoms are shown as spheres, with carbon colored cyan and hydrogen light grey, and active-site residues as a surface representation, with atoms colored depending on the structure, as indicated. Note that BAL was oriented as it should enter the active site, so that its carbonyl group is directed towards the catalytic cysteine and therefore not seen in the figure. The figure was constructed with PyMOL (https://pymol.org/).
It is then clear that an important structural feature of the four P. aeruginosa AMADHs that defines their substrate specificity is the size of the aldehyde-entrance tunnel, which is mainly defined by the size and conformation of the acidic residue sidechain and the anchor loop conformation. Asp459 and Asp457 in PA5312 and PA0219, respectively, are not a steric barrier for binding of most aminoaldehydes apart from BAL, due to their position in the active site and their sidechain being shorter than that of the glutamate. Neither is Glu446 in PA5373 because it is outside the active site, as already mentioned. On the contrary, Glu457 considerably narrows the PA4189 aldehyde-entrance tunnel, and is the main determinant for this enzyme being highly specific for ACTAL [15], which is the smallest aminoaldehyde. Regarding the anchor loop, despite its similar size and similar amino acid sequence in the four AMADHs (Fig. 7(a)), it has a very different conformation at the region that forms part of the active site, while exhibiting similar conformations in the rest of the loop (Fig. 7(b)). This finding supports the importance of the loop in the aldehyde recognition and binding. In the PA5373 crystal structure, the interactions of the Glu446 sidechain with residues of the opposing subunit (not shown), contribute to an anchor loop conformation retracted from the active site, and consequently the aldehyde tunnel in this enzyme is much more open than the one in the PA4189 crystal structure (Fig. 7(b)). In the AlphaFold model of the PA0219 structure, the anchor loop has a similar conformation to that in the crystal structure of PA5373, but this has to be confirmed when the crystal structure of this enzyme is determined. Finally, the conformation of the active-site region of the PA5312 “anchor” loop permits its aldehyde-entrance tunnel to be even more open than in PA5373, which allows PA5312 to bind the large aldehydes GluAPAL, GluABAL, and GdBAL. Fig. 8 shows a surface representation of the aldehyde-entrance tunnel that depicts its size and shape in the four enzymes.

Fig. 7 The anchor loop of the four P. aeruginosa AMADHs. (a) Amino acid sequence comparison was performed with Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/). The acidic residue is colored red and the aromatic-box residue is colored cyan, except the equivalent valine residue of PA5373, which is colored purple. (b) Structural superposition showing the mainchain atoms as cartoon and the sidechains of the acidic and aromatic residues as sticks. To better indicate the position of these residues in the active site, the catalytic cysteine sidechain is shown as a reference.
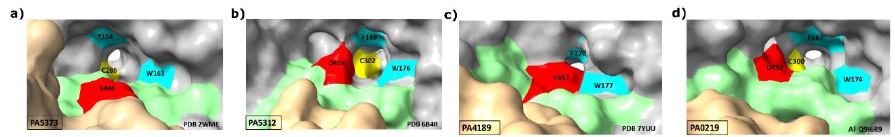
Fig. 8 Surface representation of the aldehyde-entrance tunnel of the four P. aeruginosa AMADHs. Atoms are colored light grey, except those of the anchor loop, which are green, the acidic residues red, the aromatic residues cyan, and the catalytic cysteine yellow. The atoms of the opposing subunit that interact with the anchor loop, are colored tan. The two aromatic residues shown are at the base and top of aldehyde-entrance tunnel. The third aromatic-box residue in PA5312, PA4189, and PA0219 cannot be seen because of the figure orientation. The catalytic cysteine is at the end of the aldehyde-entrance tunnel, where it connects with the nucleotide-entrance tunnel. This residue is not seen in the PA4189 figure because this enzyme has a very narrow aldehyde-entrance tunnel.
Conclusions
Substrate specificity has two requirements: the enzyme has to bind well its physiological substrate and at the same time discriminate against other chemically related compounds that it may encounter in its environment. Together, the kinetic and structural results described and discussed in this work show that the four P. aeruginosa AMADHs fulfill these requirements. The process of natural selection has worked to adjust their substrate specificity to the function that these enzymes play in the organism’s metabolism by fine-tuning their active-site structural and chemical features to the structural and chemical features of their substrates.











 nova página do texto(beta)
nova página do texto(beta)



