Introduction
The human respiratory system is responsible for the gas exchange (intake of air containing oxygen and excretion of carbon dioxide) between the external and internal environments within living organisms; specifically, in the medical field, this is defined as breathing or respiration [1,2]. In humans, the respiratory system is anatomically divided, in upper and lower respiratory tract [2]. The upper respiratory tract includes nasal cavities and an oral cavity (entry points of nose and mouth, respectively), pharynx, tongue, epiglottis, soft and hard palates and the larynx; the lower tract includes the trachea, bronchi (main stem, lobar and segmental), and lungs (bronchioles, alveolar ducts, sacs and alveoli) (Fig. 1) [3].
Before 2019, lower respiratory tract infections (LRTIs) were considered the fourth leading cause of death worldwide [4], and the ninth in Mexico [5]. This scenario has changed dramatically in the last year, where Covid-19 has caused a considerable increase in excess mortality worldwide [6]. In the United States alone, the Centers for Disease Control and Prevention (CDC) declared Covid-19 as their third leading cause of death, surpassed only by heart disease and cancer [7]. However, besides SARS-CoV-2 (the virus accountable for Covid-19), there are other important pathogens that cause LRTIs, such as bacteria and other viruses, and fungi to a lesser extent [8,9]. Viruses are challenging pathogens due to their relative high rate of mutation [10]; as a consequence, their infections are more difficult to fight [11]. This is reflected in the relative lack of effective therapies to combat viral respiratory infections, compared to the range of antibiotics known to treat bacterial infections [12].
The first record on the isolation of a respiratory infection-related virus was influenza A virus in 1933 [13]. Since then, more viruses and their variants have been related to respiratory diseases. LRTIs-causing viruses are found into 6 main viral families: Paramyxoviridae, Orthomyxoviridae, Coronaviridae, Adenoviridae, Picornaviridae and Parvoviridae[14].
General characteristics of the Paramyxoviridae family
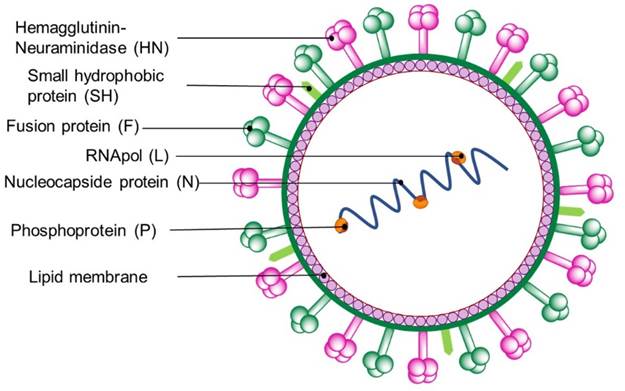
Many viruses of the Paramyxoviridae family usually display spherical shapes, and measure ca. 150 nm in diameter (there are some examples featuring 300-500 nm diameter) [15]. Paramyxoviridae exemplars have a lipid envelope surrounding the nucleocapsid and contain two or three spike-shaped transmembrane glycoproteins [16]. These proteins include: a hemagglutinin-neuraminidase spike (HN), responsible for virus attachment, a fusion protein (to induce membrane fusion) [17], and a small hydrophobic protein. Viruses of this family contain genomes consisting of 13-19 kb ssRNAs (Fig. 2). The most common respiratory illness-causing Paramyxoviridae viruses are respiratory syncytial virus (RSV) and parainfluenza viruses (PIV) (both classified within the Paramyxovirinae subfamily) and the human metapneumovirus (hMPV), categorized within Pneumovirinae subfamily.
Respiratory syncytial virus (RSV)
The respiratory syncytial virus (RSV) was discovered in 1957 from lung secretions of infants suffering from an LRTI [18]. A cytopathogenic agent, similar to another found in ill chimpanzees with common cold, was revealed as the cause [19]. RSV’s genome display ca.15.2 kb and 10 genes encoding 11 proteins [20]. A section of the viral proteome yields proteins NS1 and NS2 (both used to evade host immune defenses), and the matrix protein (M), which serves as the internal envelope [21]. Small hydrophobic (SH), fusion (F), and binding (G) proteins are surface proteins, involved in the virus entry and main targets of the immune response from host. RSV is one of the leading causes of bronchiolitis in children and is also a cause of pneumonia in premature infants and children with risk factors, such as heart or lung diseases [22]. RSV infections have also been found in adults, being elderly and immunosuppressed people featuring higher risk [20a,23]. At the time of writing this article, no RSV vaccine is yet available [24,25].
There is a single option for preventing severe RSV-caused infections, which is palivizumab (Synagis®)-based therapy [25]. Palivizumab is a monoclonal antibody against RSV’s F protein and is recommended for both prevention and treatment of infants at high risk and complications related to RSV [26]. In clinical settings, most therapeutic approaches involve the use of ribavirin (1) , nucleoside analogue approved by the FDA as the single option for the treatment of RSV and other RNA and DNA viruses infections [27]. Ribavirin displays strong inhibitory activity of RNA polymerase (RNApol) [28]. Another nucleoside analogue that has shown activity in RSV’s RNApol is lumicitabine (ALS-8176, 2), which is the prodrug of ALS-8112 (3)[29]. Examples of non-nucleosidic RNApol inhibitors are YM-53403 (4)[30], AZ-27 (5) and BRD9101 (6)[31]. A variety of promising compounds, targeting the RSV’s fusion protein (F), involved in diverse phases of clinical trials, are presatovir (GS-5806, 7)[32], sisunatovir (RV-521, 8)[33] and ziresovir (AK-0529, 9)[34].
Human parainfluenza virus (hPIV)
The human parainfluenza virus (hPIV) was isolated, by the first time in 1954. Classified within Paramyxovirinae sub-family, hPIV exemplars fall into two genus: Rubulavirus (PIV types II and IV) and Respirovirus (PIV types I and III) [35]. The envelope of hPIVs features two glycoproteins: the multifunctional hemagglutinin-neuraminidase (HN), that binds specific receptors on the target cell and removes sialic acid from receptor molecules, thus improving the spread; and the fusion protein (F), involved in the union of the viral envelope with the cell membrane (concomitantly aided by HN silidases) [36].
At the end of the 20th century, almost 30% of hospitalized children with respiratory tract infections were attributed to PIV of types I-III [37]. Although hPIV infections are most common on the upper respiratory tract (e. g. causing colds), LRTIs based on hPIV cause severe conditions such as bronchopneumonia and bronchiolitis [38]. Although there is no approved vaccine to prevent parainfluenza virus infections in humans, treatment consists of palliatives, mainly [39]. However, compounds with structural resemblance to N-acetylneuraminic acid (Neu5Ac, 10), have demonstrated efficacy both in vitro and in vivo. For example, Neu5Ac2En (11), Zanamivir (12), BCX-2798 (13) and BCX-2855 (14) are indicated as promising inhibitors of the HN protein [40].
Human metapneumovirus (hMPV)
In 2001, van de Hoogen discovered a new respiratory virus causing RSV-like symptoms [41]. The name human metapneumovirus (hMPV) was based on its biological properties and limited sequence information; however, elucidation of its genomic sequence led to the classification of hMPV as a member of the Paramyxoviridae family [42].
hMPV is pleomorphic, measuring 150-300 nm of diameter and has a single-stranded RNA genome (ca. 13 kb encoding 9 proteins) [43]. Phylogenetic analysis of hMPV revealed two main groups (A and B), divided in four genetic lineages (A1, A2, B1, and B2) [42]. hMPV entry into the host cell is rather unusual, as fusion is achieved by fusion protein (F) without intermediacy of glycoprotein (G); F is synthesized from a precursor protein (F0), cleaved by a cellular protease into two sub-units (F1 and F2) producing a hydrophobic fusion peptide (FP) that inserts into the host membrane [44]. FP is then considered a relevant therapeutic target. Ribavirin (1), heparin (15) and sulfated sialic lipid NMSO3 (16), were found to inhibit replication of serotypes 1 and 2 of hMPV [45]. An in vitro and in vivo studio of adeninic derivative IC-87114 (17), in combination with immunostimulant poly I:C, revealed replication inhibition of hMPV on human bronchial epithelial cells [46]. A recent communication revealed interesting in vitro activity against enterovirus and metapneumoviruses by using TriplatinNC (18)[47], through the formation of adducts with cell-surface glycosaminoglycans, thus blocking the viral entry. An in vitro study suggested ingavirin (19) has the potential to reduce viral replication of hMPV, 24 hours after infection [48].
General characteristics of Orthomyxoviridae family
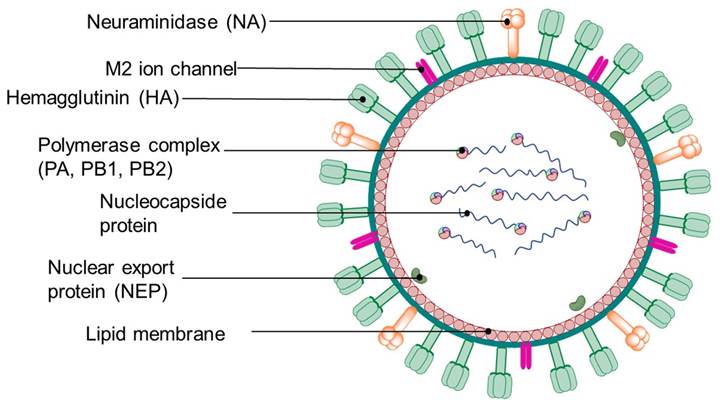
Viruses of this family are spherical or pleomorphic, with diameters of 80-120 nm and have an RNA genome of 10-14.6 kb; they also feature spike projections consisting of hemagglutinin (HA) and neuraminidase (NA) glycoproteins [49]. Spike glycoproteins have a length of 10-14 nm and a diameter of 4-6 nm (Fig. 3). Influenza virus (IV) is the most common virus that causes respiratory diseases [50].
Influenza virus (IV)
There are three influenza viruses (IVs) known to be infectious to humans: A, B and C [51], which have been the main cause behind some of the most important pandemics in the recent history of mankind [52,53]. IV’s RNA genome consists of eight segments (seven for influenza C virus) that encode 9 to 11 proteins, including surface proteins hemagglutinin (HA) and neuraminidase (NA), as well as three RNA polymerase subunits (PA, PB1 and PB2). IVs also feature two nonstructural proteins (NS1 and NEP) and two matrix proteins (M1 and M2) [54]. According to the World Health Organization (WHO), effective and safe vaccines are currently available for the prevention of influenza [55]; however, their specificity is limited due to the diversity and genetic variability of the strains, thus, vaccines are often inactive on a certain circulating subtype, or become ineffective in a short period of time [56].
IV’s surface proteins (HA and NA) are the main therapeutic targets. Antiviral alternatives against influenza viruses prevent viral entry and fusion [57]. Currently, there are four FDA-approved antivirals against influenza, three of them work as NA inhibitors: oseltamivir (Tamiflu®, 20), zanamivir (Relenza®, 12) and peramivir (Rapivab®, 21); while baloxavir (Xoflluza®, 22), acts as an endonuclease inhibitor [58].
In addition, there are currently in vitro, in vivo, and clinical studies of other promising anti-influenza compounds; some are based on oseltamivir: NA inhibitors such Laninamivir octanoate (23)[50,57,59], HA protein inhibitors BMY-27709 (24) and MBX-2546 (25), and viral replication inhibitors favipiravir (26) and pimodivir (27)[60]. Some compounds that have shown in vitro antiviral activity (as M2 channel inhibitors) are copper based compounds 28 and 29, and cobalt compound (30)[61]. Amantadine (31), previously considered as a useful M2 ion channel inhibitor, was found ineffective against influenza B viruses. As of today, influenza type A virus strains are essentially resistant to adamantanes, so they are no longer used for the treatment of IVs infection [62].
General characteristics of Coronaviridae family
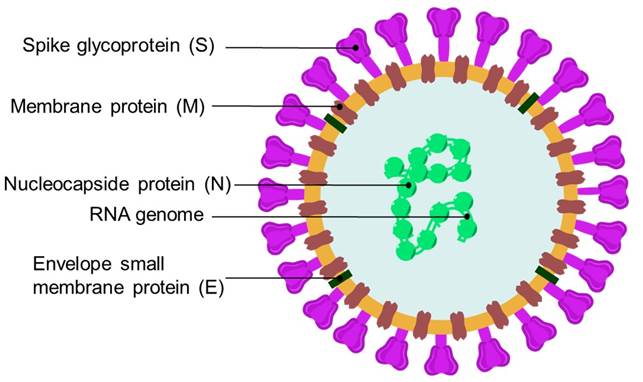
Coronaviridae family members are spherical, with 60-220 nm of diameter and 26-32 kb of positive sense RNA-based genome [63]. Coronaviridae viral particles are decorated with large club- or petal-shaped surface protein projections, known as spike (S), which adhere to host cell receptors [64]. The spike protein (S) is a large glycoprotein that mediates receptor-binding and membrane fusion (Fig. 4) [65]. In addition, Coronaviridae viruses feature a main proteinase (Mpro, also called 3CLpro) that mediates the activities of coronavirus replication process, thus making it an appealing target [66].
Human coronavirus (HCoV)
Between 1931 and 1935, cases of respiratory diseases caused by a pathogen in chicks, resembling infectious laryngotracheitis, were documented [67]. In 1936, it was proven the cause of these cases and outbreaks in hatcheries were attributed by a novel, “filterable” pathogen; early term to refer what later was known as viruses.[68] In 1965, Tyrell led an effort for the cultivation and description of common-cold virus in organ cultures (including those from humans) and revealed a virus “unrelated to any other known virus of the human respiratory tract”; this virus was isolated from a young male with a typical common cold [69]. Several authors point out to this work as the first isolation and description of a coronavirus [70]. Shortly after, in 1968, a group of 8 virologists (lead by Tyrell) wrote to Nature a letter claiming the finding of a new viral family named coronavirus, sustained by early electron microscopy images of the virus featuring “rounded profile and a characteristic “fringe” of projections which are rounded, or petal shaped” [71].
It was until 1975, when the viruses above described were accepted as a new genus called Coronavirus; and Human Coronavirus (HCoV) was among the species known (for many years the suggested acronym was HCV, now obsolete; not to be confused with the actual hepatitis C virus acronym) [72], belonging to the Coronaviridae family [73]. These viruses mutate with ease and can genetically recombine when two different strains simultaneously infect the same cell [74,75]. Coronaviruses are classified into three main groups [76]: I - alphacoronavirus, II - betacoronavirus, III - delta coronavirus (which comprises avian CoVs only) [77]. Viruses HCoV-229E and HCoV-NL63 (from group I), besides HCoV-OC43 and HCoV-HKU1 (from group II) are causative of URTIs (such as common cold); however, they have been associated with pneumonia in children, immunosuppressed patients, elderly people, and military personnel, causing serious and even fatal clinical conditions [78].
There is no particular drug therapy to counter this family of viruses. Currently, several compounds (32-47) have been tested as possible candidates for the treatment of coronavirus infections; they target the spike protein and the replication-transcription complex of the virus, thus inflicting damage on the fusion and viral replication steps [79].
Middle East Respiratory Syndrome Coronavirus (MERS-CoV)
Middle East Respiratory Syndrome Coronavirus (MERS-CoV or MERS) belongs to the genre Betacoronavirus [80]. This virus shares genetic similarity with other coronavirus present in bats, and has been detected in camels in North Africa and the Arabian Peninsula [81]. MERS caused severe acute respiratory illness in 2012 in the Arabian Peninsula, with a mortality rate of 35 % [82]. Until today, there are no vaccines nor specific antiviral treatment to combat this disease; treatment is limited to symptomatologic care [82b, 83]. Some compounds have been proposed as inhibition agents due in vitro activity against MERS and could serve as leads to develop effective drugs. Some compounds with antiviral activity against MERS are K-11777 (41), leucinamide N3 (44), leucinamide analog (45), and E-64-D (48); these compounds are protease inhibitors preventing the entry of viral genetic material into the target cell [79b,84]. Another class of antivirals (nucleoside analogs) such as mizorbine (43) and remdesivir (46), nitazoxanide (51) display broad-spectrum in vitro antiviral activity against MERS and other viruses (including RSV, PIV, influenza virus and other coronaviruses) [85]. Mycophenolic acid (Myfortic®, 52), used to prevent rejection in organ transplantation, and some flavonoids such as bavachinin (53) and quercetin (54), were tested against MERS-CoV, although their inhibition mechanism has not yet been clarified [86].
Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV)
Severe Acute Respiratory Syndrome coronavirus (SARS-CoV or SARS) emerged in southern China in 2002 and spread to about 30 countries in Asia, Europe, and North America [87]. 8098 cases were identified worldwide, with 744 deaths and a mortality rate of 9.6 % [88]. SARS genome consists of a RNA molecule with 29.7 kb, coding for 5 proteins: replicase proteins (1a and 1b), spike protein (S), envelope protein (E), membrane protein (M) and nucleocapsid protein (N) [89]. As SARS-CoV caused severe illness and threatened with a widespread pandemic, in less than a year, diverse vaccines were developed to prevent such scenario [90]. Prophylactics included inactivated virus vaccines, recombinant virus proteins, or recombinant vaccines expressing SARS-CoV S protein. The main target in the search for an effective treatment against SARS-CoV has been the Mpro. Diverse peptides with Michael acceptor moieties such as leucinamide (44), Rupintrivir (AG-7088, 49) and KZ-7088 (50), or aldehydic derivatives such as TG-02005221 (55) were effective against Mpro in vitro, as highlighted in some reviews [91]. Cinanserin (56) and analogs are another class of Mpro inhibitors [92]. Other drugs with promising antiviral activity in vitro against SARS-CoV have been identified by high throughput screening of chemical libraries [93]. Among these, MP-576 (57) featured activity against Mpro, while VE-607 (58) inhibited viral entry, presumably by impeding binding to the ACE2 receptor. FK506 (37) showed activity against Nsp1 protein of SARS-CoV [79a].
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2)
In December 2019, a new coronavirus emerged in Wuhan, China, causing a worldwide health emergency leading to millions of deaths globally [94]. The WHO declared the worldwide health scenario caused by Covid-19, formal name of the disease caused by the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) [95], as a pandemic on March 11, 2020 [96]. SARS-CoV-2 virus is significantly related to a bat coronavirus and showing a ca. 76 % of amino acid sequence identity to the severe acute respiratory syndrome coronavirus (SARS-CoV) [97]. The SARS-CoV-2 genome size ranges from 29.8 to 29.9 kb, where more than two thirds of the genome encode for polyproteins, while one third consists of genes that encode for structural proteins including spike (S), envelope (E), membrane (M) and nucleocapsid (N) [98]. After 183 million confirmed cases and currently 3.98 million deaths [99], it has become one of the worst pandemics in the history of mankind [100]. Until June of 2020, several variants have emerged around the world, including those classified as variants of interest and variants of concern [101]. This has led to global efforts to implement vaccination campaigns [102], as well as drug repurposing studies in antiviral therapies directed towards SARS-CoV-2. Small molecule reuse has been proposed for the treatment of Covid-19, based on their previous efficacy against SARS-CoV and MERS-CoV in vitro[103]. Some drugs exhibited activity on the main therapeutic targets of SARS-CoV-2 [104]. Chloroquine (CQ, 32), hydroxychloroquine (HCQ, 33) and baricitinib (Olumiant®, 59), have been involved in clinical trials against Covid-19. 32 and 33 are antimalarian drugs that disrupt endosomal pH, but they resulted non-effective in treating Covid-19 [105]. Both 32 and 33 have been extremely controversial as their use was clearly rejected by the FDA since there was no therapeutical significance on decreasing mortality rates from Covid-19; in fact both 32 and 33 caused adverse, undesirable events in patients that received these drugs to counter the disease [106]. Baricitinib features anti-cytokine activity and in co-administration with remdesivir has shown accelerated improvement of clinical patients with Covid-19 [107]. To prevent the entry of SARS-CoV-2 in host cells, drug therapy that hampers the interaction between the SARS-CoV-2 S protein and the human ACE2 receptor is desired [108]. The use of angiotensin II receptor antagonists, such as Telmisartan (Micardis®, 60) and losartan (Cozaar®, 61) has been proposed as a strategy to reduce ACE2 expression, and hence, viral entry; however, their putative beneficial effects in clinical trials may not be related to ACE2 expression levels [109]. Many peptides and peptide-based antivirals, such as ATN-161 (62), have been identified as potential blockers of the interaction of the viral S protein with the ACE2 receptors [110, 111]. Finally, TMPRSS2 serine protease is also involved in membrane binding and the entry of viral material in host cells. Nafamostat (63) and camostat mesylate (64) are two compounds with inhibitory activity against TMPRSS2 [112]. Recently auranofin analogs such as 65 have been reported with in vitro activity inhibiting Spike-ACE2 interaction, comparable to disulfiram (66), both also targeting papain-like protease (PLpro) in SARS-CoV and SARS-CoV-2 [113].
The two cysteine proteases Mpro and PLpro, responsible for the polyprotein processing and generation of new virions, have become major targets for the development of drug therapies against SARS-CoV-2 [114]. Many drugs have been tested for inhibitory activity of Mpro: lopinavir (39) in combination with ritonavir (40)[115], darunavir (67) and cobicistat (68)[116]. Bepridil (69) pimozide (70) and ebastine (71) have been tested in silico and in vitro, showing promising results against Mpro of SARS-CoV-2 [117]. Other compounds with remarkable activity on Mpro are dalcetrapib (72)[118], GC-376 (73)[119], boceprevir (74) and calpain inhibitors 75 and 76[120], involved in various pathologies such as Alzheimer and cataract formation [121]. Recently, Pfizer reported that PF-07321332 (77), a prodrug of PF-07304814 (78), have entered to phase 1 clinical trials for the treatment of SARS-CoV-2 as Mpro inhibitors [122]. Another molecule is PF-00835231 (79), which is also active against Mpro[123].
RNA-dependent RNA polymerase (RdRp) is the most important component of the genome replication and transcription complex and has been considered as an important target for nucleotide-based antivirals [124]. Some compounds such as molnupiravir (EIDD-2801, 80) and its prodrug EIDD-1931 (81)[125], tenofovir (82)[126], IDX-184 (83)[127], and combinations of sofosbuvir (84) and ledipasvir (85)[128], or daclatasvir (86)[129] have been tested with promising results. Recently, Merck has made a request for the emergency use of 80 to treat Covid-19 in the US, due to the favorable results of this drug in current clinical trials [130]. Nowadays, the only drug accepted by the FDA to fight Covid-19, is remdesivir (Veklury®, 46) [131]. 46 is a monophosphoramide developed by the pharmaceutical company Gilead Sciences, Inc. 46 is a prodrug that releases intracellularly GS-441524 (47)[132], whose triphosphate inhibits viral RNA polymerase through its attachment and subsequent termination of replication in the chain of Viral RNA [133].
General characteristics of Adenoviridae family
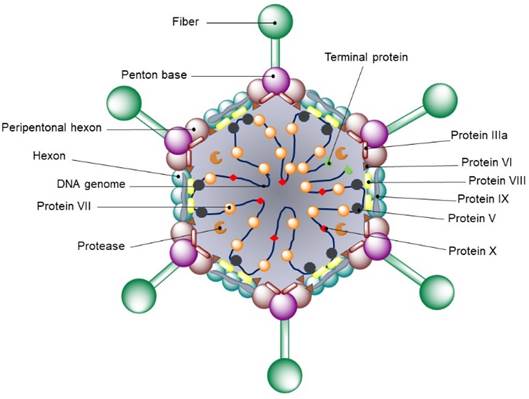
Viruses within Adenoviridae family are 70-90 nm in diameter and do not have an envelope [134]. The icosahedral capsid consists of 240 capsomeres of 8 to 10 nm in diameter and 12 vortex capsomeres (penton bases). Each capsomer features a fiber projecting from the surface of the virion, resulting on the characteristic morphology of this family (Fig. 5). Additional components of the outer layer are polypeptides IIIa, IX and VI, which are in contact with other coating proteins called hexons, making up a continuous protein layer. Adenovirus genome consists of a single DNA molecule ranging from 26-45 kb and contains an inverted terminal repeat (IRR). Genome’s central component is conserved throughout the family, while the two ends show variations in length and genetic content [135].
Human adenovirus (HAdV)
Adenoviruses (HAdVs) are a very common cause of respiratory infections in people of all ages [136]. HAdVs structure is a medium-sized unenveloped icosahedra (90-100 nm), with double-stranded DNA [137]. More than 50 adenovirus serotypes can cause infections in humans.
Currently, there is no specific antiviral drug approved by the FDA for the treatment of adenovirus [138]. Clinical management of adenovirus-caused infections are based in palliative care. However, some molecules feature inhibitory activity as they prevent viral DNA replication. Cidofovir (87) has shown increased activity compared to ribavirin and has been used in serious cases of immunocompromised individuals [139]. However, side-effects have been observed with this drug due to kidney toxicity; therefore, its use is limited (for other illness) and currently, has not been approved by the FDA for adenovirus infections [14]. Other promising drugs such as brincidofovir (88) and 9- (3-hydroxy-2- phosphonylmethoxypropyl) adenine (HPMPA, 89), have shown in vitro and in vivo efficacy for some adenovirus serotypes, and derivatives from niclosamide (90) (discontinued in the US and other countries for treating parasitic tapeworm infections) [140], such as JMX-0312 (91) and JMX-0281 (92) exhibit antiviral activity in vitro. Unfortunately, some of these compounds have not been evaluated beyond in vitro studies due to their side effects; their use is still under debate [141].
An application of adenoviruses in vaccine development is their use as vectors. Adenoviruses genetic material is easy to manipulate and are capable to trigger T and B cells response [142]. Several vaccines are currently in development against viruses such as ebola [143], human immunodeficiency virus (HIV) [144], and SARS-CoV-2 [145]. Interestingly, some vaccines use adenovirus serotype 5 (Ad5), and this serotype is widespread as the cause of the common cold; unfortunately, individuals previously exposed to Ad5 are prone to have high levels of neutralizing antibodies, resulting in immunological response against the vaccine [146]. Alternative strategies use less-common adenoviruses, such as adenovirus serotype 26 and 35 (Ad26, Ad35); anti-SARS-CoV-2 vaccines such as Sputnik V (Gamaleya) and Ad26.COV2.S (Johnson&Johnson, Beth Israel Lahey Health, Janssen) feature Ad26 vector. Cellid and LG Chem are developing an anti-SARS-CoV-2 vaccine using Ad5 and Ad35 as vectors [147]. Vaxart and ImmunityBIO vaccine efforts feature Ad5, the strain used for Convidecia and Sputnik V vaccines (CanSinoBIO and Gamaleya, respectively). RSV virus are also found in anti-SARS-CoV-2 vaccine research, as Meissa Vaccines is developing a spray alternative which is now in Phase I trials.
General characteristics of Picornaviridae family
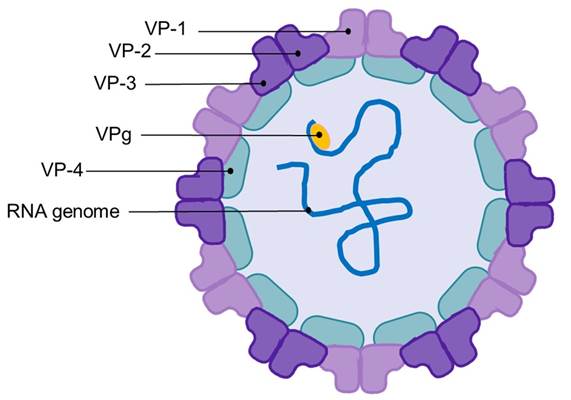
Picornaviridae family members are considered as the most common source for human viral diseases [148]. Named picornavirus due to their small size (22-30 nm) and relatively small genome (comprising a single RNA strand of 7.0-8.8 kb), its viral structure is an icosahedral capsid (Fig. 6) [149]. Picornavirus capsid is composed of 60 protein units (classified as VP-1, VP-2 and VP-3), and, in most picornaviruses, an internal protein (VP-4). Unlike other viruses, picornavirus lacks external projections; however, picornaviruses feature a genome-linked viral protein (VPg), which serves as a primer during RNA synthesis.
Rhinovirus
Human rhinovirus is the main and most common cause of the common cold and is the best-known member of the Picornaviridae family [150]. This viral subclass occasionally causes severe cases of LRTIs such as bronchiolitis, pneumonia, and exacerbations of chronic lung disease [151]. Rhinovirus are the smallest in comparison to other infectious viruses (approximately 30 nm) and its genetic material consists of 7.2 kb of RNA of RNA (coding for 11 proteins) [152]. Variations in the capsid results in more than 100 serotypes of this virus. Rhinoviruses also possess non-structural proteins including a polymerase (3Dpol), two proteases (2A and 3C), and structural proteins 2B, 2C, 3A, and 3B [153].
Most rhinovirus serotypes bind to host cells through ICAM-1 cell adhesion molecules, while a few serotypes bind and enter the cell through low-density lipoprotein receptors [154]. Rhinovirus binding mechanisms are the most studied and target of antiviral therapies; for example, pleconaril (Picovir®) (93), binds to a hydrophobic pocket in the VP1 protein, thus hampering the installation of ICAM-1 [155]. FDA does not recommend the use of pleconaril due to obnoxious side effects (headache, nausea, and diarrhea) [156]. Other molecules, featuring antiviral activity against rhinovirus are vapendavir (94) and rupintrivir (49)[157]; however, to our knowledge, none of them have been approved or recommended. Other compounds that have shown in vitro activity against picornavirus, are pyrithion (95) and hinokitiol (96), whose zinc salts have shown inhibitory action on the processing of viral polyproteins [158].
General Characteristics of Parvoviridae family
Parvoviruses display icosahedral shape [159] with diameters of 23-28 nm, and smooth or pointed “finish”. Their genome consists in a ssDNA molecule (4-6.3 kb in size) encoding for 2 to 4 virion proteins (denominated VP1-VP4) (Fig. 7) [160].
Human bocavirus (hBoV)
The human bocavirus (hBoV) was discovered in 2005 and was isolated from samples obtained from children with respiratory tract infections [161]. Phylogenetic analyses revealed that hBoV is a parvovirus closely related to bovine and canine parvoviruses. hBoV feature 43 % of resemblance to canine parvovirus, thus it was included within the Parvovirinae subfamily. Since its discovery, HBoV has been reported in blood, fecal, and urinary samples [162]. There is no approved treatment for HBoV infections; therefore, symptomatic treatment is like other viral respiratory tract infections. Until 2018, cultivation of HBoV remained a challenge [163]. Therefore, implementation of studies looking for effective drug therapies to counter HBoV, are limited; no results have been published regarding to such efforts [164].
Conclusion
It is undeniable the impetus to find effective drugs against respiratory infections caused by viruses, especially due to the current Covid-19 pandemic, which has stimulated research on new and repurposed drugs. While using high throughput screening of existing drug libraries in search for antiviral activity, it is important to identify the molecular targets of each molecule in vitro, especially in repurposing drugs [165]. Although there is uncertainty in research that aims to develop new antiviral agents, due to the mutagenic nature of viruses, efforts must be boosted in all areas to develop effective antiviral therapies. Antiviral research must catch the attention of the scientific community just at the same level as vaccines, leading towards multivectorial efforts to combat viruses.











 nueva página del texto (beta)
nueva página del texto (beta)