Introduction
On November 12, 2019, the first case of infection of a novel coronavirus (COVID-19) was identified at Wuhan, the capital of Hubei province in China. [1] Regardless of the origin of this virus, which is uncertain, it has produced a pandemic that has been the cause of human deaths worldwide. Governments have attempted to control the pandemic by requesting that the population maintain a healthy distance and stay at home. This produces collateral damage, as it affects the economy of the population. Social inequality in Mexico is particularly shameful. Fifty percent of the population lives in poverty and survives on what they can earn each day. For these people, staying at home has many repercussions, making it difficult to stop the pandemic solely with these actions (social distancing and remaining at home). Therefore, it is very important to develop the vaccine and/or find drugs to help control this infection.
The Coronaviridae family is constructed from enveloped viruses with a positive-strand RNA genome. [2,3] These coronaviruses mostly contain four structural proteins: spike protein, envelope protein, membrane protein, and nucleocapsid protein. [4] Spike proteins are relevant during virus infection, as they promote host attachment and virus-cell membrane fusion. [5] COVID-19 is a coronavirus that belongs to this family. Several compounds have been investigated for the purpose of developing antiviral drugs or a vaccine against human COVID-19 infection, but an effective and definitive antiviral strategy to halt this emergency has still not been devised.
Two drugs that have been used for this purpose are cloroquine and hydroxychloroquine. First synthetized in 1934, cloroquine is a medicine for the treatment and prevention of malaria and autoimmune conditions (rheumatoid arthritis and lupus erythematosus for example). Hydroxychloroquine was produced later in 1955, and was reported to be as effective as cloroquine against malaria, but less toxic. It was later stated that based on its physicochemical properties, hydroxychloroquine was much less active than cloroquine. Both were studied for the treatment of SARS-CoV, which is the virus responsible for the SARS coronavirus epidemic that we suffered in 2002. [6-9]
Up until now, it has not been clear whether these drugs are effective for the treatment of the COVID-19 infection. [9-11] French studies reported that hydroxychloroquine combined with azithromycin might reduce the viral load in patients, [12,13] but other studies reported that there was insufficient evidence to verify the effectiveness of these drugs against COVID-19. [14] However, in previous studies authors suggested that the lack of effectiveness of these drugs may relate to other patient conditions, such as renal or hepatic damage. Although there are many clinical studies, the action mechanism of chloroquine and hydroxychloroquine against COVID-19 is not yet fully understood. One of the hypotheses is that both drugs can be incorporated into endosomes and lysosomes, producing an increment in the pH of the intracellular compartments. This produces dysfunction and affects the conditions required for viral infection, replication and propagation of the virus. [15]
In order to understand the action mechanism, it is important to have accurate information about any molecules that might be used in the treatment of COVID-19. In spite of all the studies reported until now, there are no theoretical studies concerning the electronic properties of these drugs. For this reason, the main objective of this report is to present electronic properties of chloroquine and hydroxychloroquine that may help elucidate the action mechanisms against this virus. Even though now it is known that these two molecules do not have positive effect on the disease produced by the virus and apparently they are not longer useful for the treatment of COVID-19, it is important to have an electronic characterization of these drugs, since they were used with success with several patients (Plaquenil ® was used in Mexico for patients with COVID-19) and they were also useful for the treatment of other illness.
Computational details
Gaussian09 was used for all electronic calculations. [16] Geometry optimizations without symmetry constraints were implemented at M062x/6-311+G(2d,p) level of theory [17-21] while applying the continuum solvation model density (SMD) with water in order to mimic polar environments. [22] Harmonic analyses were calculated to verify local minima (zero imaginary frequencies). We considered protonated states of all drugs following the available experimental evidence. PubChem configurations were used as initial geometries.
Conceptual density functional theory (CDFT) is a chemical reactivity theory founded on Density Functional Theory based concepts. [23-27] Within this theory there are response functions that help us to understand the chemicial reactivity. The response functions that we used in this investigation are the electro-donating (ω-) and electro-accepting (ω+) powers, previously reported by Gázquez et al. [28, 29] These authors defined the propensity to donate charge or ω- as follows:
whereas the propensity to accept charge or ω+ is defined as
I and A are vertical ionization energy and vertical electron affinity, respectively. Lower values of ω- imply greater capacity for donating charge. Higher values of ω+ imply greater capacity for accepting charge. In contrast to I and A, ω- and ω+ refer to charge transfers, not necessarily from one electron. This definition is based on a simple charge transfer model expressed in terms of chemical potential and hardness. These chemical descriptors have been used successfully in many different chemical systems. [30-34]
Results and discussion
Cloroquine and hydroxychloroquine represent diprotic weak bases that are protonated at physiological conditions, depending on pKa values. It was reported that these drugs should be administered in tablets in its basic form. For this reason, it is important to calculate neutral and protonated molecules. Fig. 1 reports the optimized structures of cloroquine (neutral and protonated). Bond distances between the nitrogen atom of pyridine and a carbon atom of one methyl group are included in each case. This indirectly indicates the doubling of the molecule. Evidently, the di-protonated molecule is less folded than the neutral and the bond distance of the mono-protonated structure is the shortest. For hydroxychloroquine, the optimized structures (see Figure 2) indicate that molecules are also folded. The distance between the nitrogen atom of the pyridine and the oxygen atom is an index of the molecular angle. The mono-protonate structure has longer distance than the neutral, with the di-protonated molecule representing the shortest. These modifications to structure may relate to the action mechanism. In terms of geometry, there is no significant difference between these two drugs. Under physiological conditions, both protonated species are present and both may contribute to incrementing the pH values in the intracellular compartments. These optimized structures should be taken into account, as the geometry of the molecule is important when there is interaction with the proteins of the virus. These optimized structure are folded, and this is in agreement with previous experimental results of the X-Ray structure [35]. Figures 1 and 2 also report the Mulliken atomic charges of some atoms. In all cases, the carbon atom that is bonded to the chlorine is positive (almost +1), whereas the chlorine atom is slightly negative. The oxygen atom of hydroxychloroquine has a negative atomic charge, as anticipated.
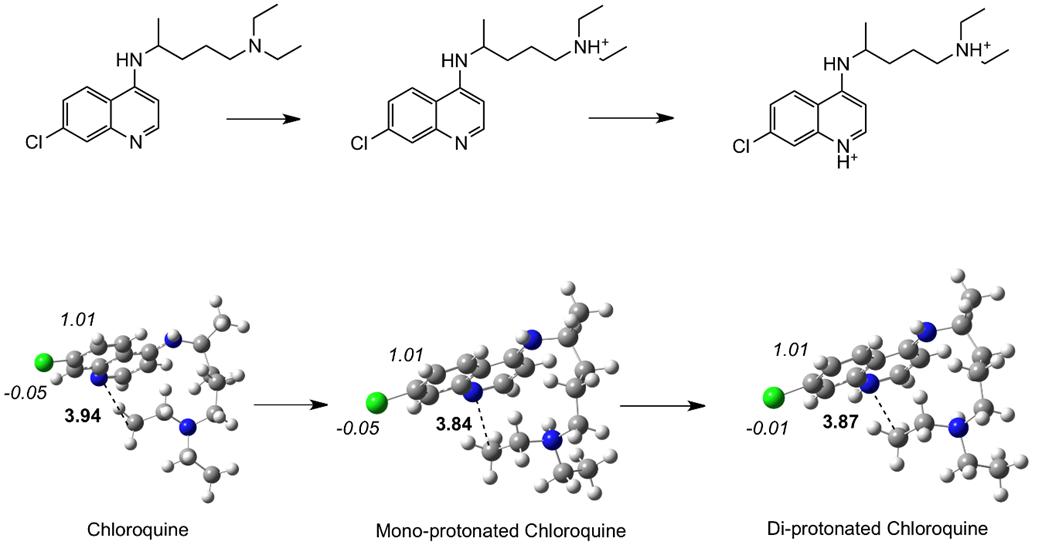
Fig. 1 Schematic representation and optimized structures of cloroquine, neutral and protonated. N-CH3 bond distances in Å (in bold) and Mulliken atomic charges (in italics) are included.
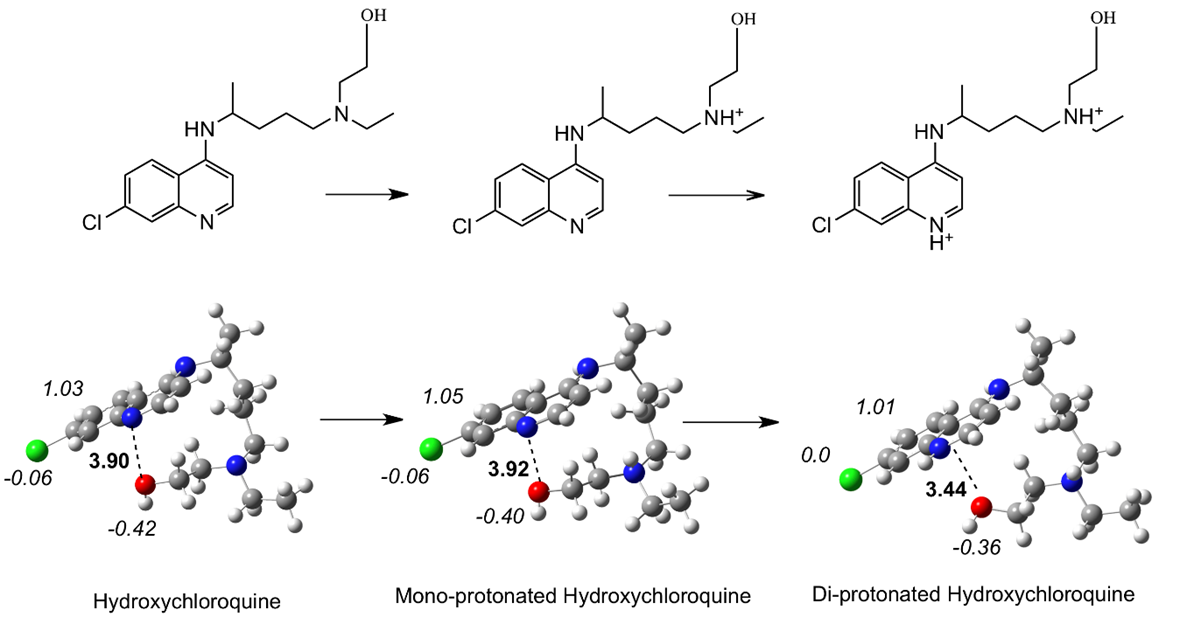
Fig. 2 Schematic representation and optimized structures of hydroxychloroquine, neutral and protonated. N-O bond distances in Å (in bold) and Mulliken atomic charges (in italics) are reported.
As the basic forms are used as drugs against diseases, it is important to analyze the di-protonated species in more detail. Fig. 3 reports the molecular frontier orbitals for the di-protonated molecules.
Cloroquine and hydroxychloroquine have practically the same molecular frontier orbitals. All molecular orbitals are located at the benzene and pyridine rings and the rest of the atoms do not participate. These orbitals are π-bonding orbitals. There is also a p non-bonding orbital of the chlorine atom and there are π-antibonding orbitals between chlorine and carbon atoms. In the case of hydroxychloroquine, the HOMO-3 orbital forms a π-bond above the region where the oxygen atom is placed. This orbital is different from the corresponding orbital of cloroquine. However, HOMO and LUMO are similar for both molecules. The electrons located in these orbitals can be said to be the active electrons, which may participate in charge transfer processes. Benzene and pyridine rings and chlorine atoms will accept electrons and the electrons will be ionized from the π-bonding orbitals of the rings.
Vertical ionization energy and vertical electron affinity were obtained for all the molecules and are reported in Table 1. As expected, protonated species have higher ionization energies and higher electron affinities than non-protonated molecules. The ionization energy reveals that it is energetically costly to donate electrons. The electron affinity indicates that it is energetically favorable to accept electrons, so that these molecules are oxidants. For comparison, we included the values for two oxidant free radical molecules in Table 1. Di-protonated molecules have higher electron affinity than NO, as they are better electron acceptors. This is important as these molecules may oxidize other molecules, such as proteins and DNA-nitrogen bases. Apparently, the oxidative capacity of these molecules is high, a factor which may affect the toxicity or effectiveness of these molecules.
Table 1 Vertical ionization energy and vertical electron affinity (IE and EA in eV) are reported. Electrodonating and electroaccepting powers (ω- and ω+ respectively) are also included.
| Molecule | IE | EA | ω- | ω+ |
|---|---|---|---|---|
| Cloroquine | 5.93 | 1.83 | 5.88 | 1.99 |
| [Cloroquine-H]+1 | 5.99 | 1.89 | 6.01 | 2.07 |
| [Cloroquine-2H]+2 | 6-77 | 2.58 | 7.81 | 3.15 |
| Hydroxychloroquine | 5.91 | 1.81 | 5.83 | 1.97 |
| [Hydroxychloroquine-H]+1 | 5.98 | 1.87 | 5.96 | 2.04 |
| [Hydroxychloroquine-2H]+2 | 6.75 | 2.55 | 7.74 | 3.09 |
| NO | 6.32 | 1.88 | 6.11 | 2.01 |
| OCH3 | 8.30 | 4.75 | 15.46 | 8.94 |
Table 1 also reports the electrodonating and electroaccepting powers of the molecules under study. These quantities refer to the partial charge transfer processes. Both quantities are increasing according to the protonation. Non-protonated molecules are better electron donors and worse electron acceptors than protonated species.
Conclusion
Expressed simply, the hypothesis about the action mechanism of these molecules indicates that they increment the basicity of the environment, thus curtailing the reproduction of the virus. This is clear from the pKa values, as under physiological conditions these molecules are mono and di-protonated. However, these molecules may also interact by transferring electrons or oxidizing other molecules of the organism, and may also oxidize proteins in the virus. The idea that emerges from this study is that not only the acid-base equilibrium is important. Besides this, the reduction-oxidation equilibrium may explain the toxicity or the effectiveness of these drugs. What is certain is that these drugs may contribute to increased oxidative stress in the body. Being a molecule able to oxidize other molecules is similar to yin-yang; a dualism that describes contrary forces, as oxidation may contribute to attacking the virus, but also contributes to increasing oxidative stress. These results should help improve understanding of the action mechanisms.











 nueva página del texto (beta)
nueva página del texto (beta)



