Introduction
Polycyclic aromatic hydrocarbons (PAHs) are well known contaminants mostly originating from natural and anthropogenic combustion processes. The US Environmental Protection Agency (EPA) have catalogued 16 PAHs as priority contaminants to their toxicity and have mutagenic or carcinogenic effects in organisms [1]. In addition to being recalcitrant and persistent in the environment [2].
PAHs in marine environments are likely to increase due to anthropogenic activities in the coastal zones, such as sewage discharges, marine traffic of ships, fuel loading and unloading and this represents a long-term environmental risk. For instance, considerable amounts of PAHs have been reported in marine sediments of several points of the Coast of Baja California, including Todos Santos Bay and Rosarito Port area in Baja California, Mexico, and high molecular weight (HMW) PAHs, as phenanthrene and pyrene, were the most abundant [3,4].
The use of native hydrocarbonoclastic marine bacteria is a feasible alternative to reduce PAHs from the marine environment. Usually, the presence of PAHs pollutants promotes the growth in these kinds of bacteria through the activation of integrated specialized machinery that consists of an enzymatic battery and mechanisms such as chemotaxis, biofilm formation, biosurfactant production, among others, which facilitate the biodegradation process. Many marine members of Bacteria domain, like Pseudomonas, Acinetobacter, Ochrobactrum, Rhodococcus, Kocuria, Arthrobacter, Micrococcus, among others, have been reported to degrade PAHs [5-7].
On the other hand, the isolation and typing of indigenous bacterial strains are important tasks for the selection of the microbial community suitable for bioremediation purposes and low the risk to human health and other wild communities. Identification of specific strains also facilitates the study of the molecular mechanism of PAHs degradation applying the emerging technology "omics". For instance, the bacterial identification/classification method per excellence has been the 16S rRNA gene sequencing PCR-based, nevertheless, is costly and time consuming and is important the selecting the accurate region gene to sequence to distinguish closely related bacterial species [8].
The molecular identification and classification of microorganisms using proteomic profiles provided by matrix-assisted laser desorption/ionization (MALDI) coupled to time-of-flight mass spectrometry (TOF MS) has been widely used over the last two decades in clinical, environmental and food microbiology [9]. Through analysis by MS of intact-cell or partial bacterial cell lysis, abundant proteins (principally ribosomal) are detected based on m/z value and intensity. The proteins profile of each strain can be used for taxonomic classification at the level of genus or species using spectra of reference organisms included is specialized libraries. In general, identification by MALDI is fast, inexpensive with effective great resolution to discriminate species and reliable results as more as16S rRNA gene sequence analysis. However, these advantages are still controversial and strongly dependent on the software/database used to analyze the result. Currently, five are commercially available systems [10].
This study was conducted to identify pyrene-utilizing bacteria isolated from the coastal area of Rosarito Port, B.C., Mexico by use MALDI-Biotyper (Bruker, Daltonics) and demonstrate that it is becoming an essential analytical tool in the environmental microbiology field.
Experimental
Reagents
All chemicals were of analytical reagent grade. Deionized water (18.2 Mῼ cm, Labconco); ethanol, acetonitrile (MeCN), trifluoroacetic acid (TFA), dichloromethane, hexane and chloroform, as well as the matrix for MALDI (α-cyano-4-hydroxycinnamic acid: HCCA) were purchased from Sigma-Aldrich. Twenty five PAHs analytical standard were naphthalene, 1-methylnaphthalene, 2-methylnaphthalene, biphenyl, 2-dimethylnaphthalene, acenaphthylene, acenaphthene, 3-trimethylnaphthalene, fluorene, dibenzothiophene, phenanthrene, anthracene, 1-methtylphenanthrende, fluoranthene, pyrene, benzo(a)anthracene, chrysene, benzo(b)fluoranthene, benzo(k)fluoranthene, benzo(a)pyrene, benzo(e)pyrene, perylene, benzo(g,h,i)perylene, dibenzo(a,h)anthracene, indene (1,2,3) pyrene were from UltraScientific.
Analysis of polycyclic aromatic hydrocarbons concentration in marine sediments and surface seawater samples
Extraction of PAHs from sediments. A modified method to PAHs extraction of sediments developed according to Murphy et al [11] using an accelerated extraction system by solvent Dionex ASE 350. A stainless steel cell of 34 mL was used. The method consists of four static cycles of 5 minutes at 100 °C and 1500 psi of constant pressure, with 60% replacement volume of total cell capacity, 180 seconds of purge and dichloromethane as extraction solvent. To remove interferences during the sample extraction, in-cell cleanup was used (layer of silica and basic alumina). The quality control included certified standard reference material (NIST SRM 1941b), procedure blanks and surrogate recoveries.
Extraction of PAHs from seawater. Seawater (1.8 L) samples were extracted with dichloromethane in funnel (3 X 60 mL). The extracts were concentrated and cleaned through liquid chromatography by column (10 mL syringe) packed with basic alumina and eluted in sequence with 10 mL hexane, 20 mL hexane:dichloromethane (1:1, v/v) and 10 mL dichloromethane. The quality control included fortified and procedure blank, and surrogate recoveries.
Chromatographic method to determine PAHs concentration in seawater and sediments samples. Gas chromatography coupled to mass-spectrometer detector (GC-MS) in electronic impact mode (EI) was used to analyze extracts obtained from seawater and sediments samples. The DB-5MS 30m long column (250 μm x 0.25 μm, Agilent Technologies, Inc) was used to separate analytes. The GC-MS conditions were as follows: 1μL was injected in the splitless mode 1 mL min-1. Helium was used as carrier gas at flow of 1 mL min-1. Injection port and interface detector were maintained at 280 °C. The oven program consisted of initial temperature at 70°C for 0 min to a final temperature of 325°C and a rate of 5 °C min-1, with a final hold time of 6 min, for a total run time of 60 min. PAHs were quantitated by selective ion monitoring (SIM) with standard mixture 25 analytes and p-terphenyl-d14 as internal standard [11,12].
Enrichment culture and bacterial isolation
Pyrene-utilizing bacteria were obtained from marine sediments and surface seawater samples collected at three sites in the coastal area of Rosarito Port, Baja California, Mexico (M, C and CFE sampling sites). The medium Bushnell Haas broth (BH) supplemented with 25 mg L-1 pyrene (as the sole carbon and energy source) was used for the bacterial enrichment and isolation. Briefly, for the first enrichment, 1 g of marine sediment o 1 mL of surface seawater from each sample was added to sterilized culture medium and then incubated in the dark at 25 ºC ± 2 ºC, in rotatory shaker (160 rpm), for 7 days. Ten percent of the first enrichment of each culture was transferred to the flasks with fresh media supplemented with pyrene. The enrichment and transfer processes were made three times. Bacterial isolates were obtained by inoculate 20, 50 and 100 µL of the last enrichment in Bushnell Haas agar plates previously sprayed with 25 mg L-1 of pyrene. Colonies morphologically distinct were recovered and submitted to the streaking method on BH agar plates supplemented with 25 mg L-1 pyrene to obtain pure cultures.
MALDI-TOF MS and identification of the isolate by Biotyper system
The isolates obtained from the previous step were identified by the MALDI-Biotyper system. Briefly, 500 µL of overnight cultures (18 h/37 ºC) in LB broth were centrifuged, the pellet was suspended in 1 mL of water and centrifuged again. The pellet obtained was mixed with 1 mL of ethanol (70%) centrifuged and the pellets were resuspended in equal parts of formic acid (70%) and acetonitrile. The protein extract of each isolate was obtained in triplicate, from independent cultures (biological replica) and 1 μL of extracts was deposited on a spot of polished steel target, air dried, and spot each were overlay with 1 μL of matrix solution (2.5 mg of HCCA in 250 µL of 50 % acetonitrile / 2.5% TFA v/v). Each biological replica was applied to five spots and acquired in duplicate. Samples were processed using a AutoFlex speed MALDI-TOF MS (Bruker Daltonics, Germany) equipped with a smartbeamTM-II Nd:YAG laser. Mass spectra profiles were recorded in a linear positive ion mode with instrument parameters optimized for the range of m/z 2,000 to 20,000 under control software (flexControl 3.0; Bruker Daltonics, Bremen, Germany). The acquisition spectrometer parameters were using the pre-programmed MBT.FC.par method: the voltages of ion sources 1 and 2 were set at 19.4 kV and 18.07 kV, respectively. The lens voltage was set at 7.0 kV. External mass calibration was done with a mixture of proteins contained in the Bacterial Test Standard (BTS, Bruker Daltonics) with an average mass tolerance of about 25 ppm.
After each measurement, the spectra were manually inspected using FlexAnalysis3.4 software to verify the quality. Spectra quality criteria of base peak signal-to-noise (S/N threshold of 5) ratio, minimum relative intensity threshold of 5%, base peak resolution, number of peaks, and mass range of peaks were selected according to previous reports [13]. Mass deviation within the spectra sets was analyzed. Flat-liners and spectra with peak variations (outliers) were removed from the collection, and additional measurements were carried out. Raw spectra were transferred to Biotyper Compass Explorer (Bruker Daltonics, Germany, Version 4.1.14) [14].Before starting the identification, the preprocessing of the mass spectra was performed and that included five steps: mass adjustment (Spectra Compressing by a factor of 10), smoothing (Savitsky-Golay algorithm with a frame size of 25 Da to the mass adjusted data), baseline subtraction (TopHat algorithm), normalization (Maximum) and peak picking, following the recommendations for Bacterial identification strategies by MALDI-TOF MS [15] was then assigned to its specific node on the taxonomy tree.
In order to visualize the discrimination or relationship between the mass spectra of isolate related [16], Principal Components Analysis (PCA) and the Composite Correlation Index (CCI) were performed using Biotyper Compass Explorer (Bruker Daltonics, Germany, Version 4.1.14).
Results and Discussion
Determination of PAHs in surface seawater and marine sediments
The analysis of 25 PAHs from samples of surface seawater and marine sediment of three sites from at area of coastal in Rosarito Port, B.C., Mexico (M, C and CFE) was performed using GC-MS (Table 1). It should be noted that in these sampling areas there is a discharge of fuel (M and C sites) and boat transit (CFE). The total concentration of PAHs presented similar values in all surface water samples analyzed (0.461 - 0.525 ng mL-1); whereas higher levels were found in the sample of marine sediment (74 -266 ng g-1 dry weight) indicating light PAHs pollution. The levels of PAHs reported in our study were resemble to the levels found in other regions of the world [17-19].
Table 1 PAHs concentrations values measured in surface seawater and marine sediments from Rosarito Port, B.C., Mexico.
| Compound | Sampling site | |||||
|---|---|---|---|---|---|---|
| M | C | CFS | ||||
| SW1 | S2 | SW1 | S2 | SW1 | S2 | |
| NAPHTHALENE (N) | 0.051 | L.D.L. | 0.063 | L.D.L. | 0.044 | L.D.L. |
| 1-METHYL NAPHTHALENE (1-MN) | 0.026 | L.D.L. | 0.028 | L.D.L. | 0.023 | L.D.L. |
| 2-METHYL NAPHTHALENE (2-MN) | 0.013 | L.D.L. | 0.015 | L.D.L. | 0.012 | L.D.L. |
| BIPHENYL (BPh) | 0.008 | L.D.L. | 0.009 | L.D.L. | 0.007 | L.D.L. |
| 2-DIMETHYL NAPHTHALENE (2-DMN) | 0.010 | L.D.L. | 0.010 | L.D.L. | 0.009 | L.D.L. |
| ACENAPHTHYLENE (Acy) | 0.004 | L.D.L. | 0.004 | L.D.L. | 0.004 | L.D.L. |
| ACENAPHTHENE (Ace) | 0.009 | L.D.L. | 0.010 | L.D.L. | 0.008 | L.D.L. |
| 3-TRIMETHYL NAPHTHALENE (3-TMN) | 0.008 | L.D.L. | 0.010 | L.D.L. | 0.009 | L.D.L. |
| FLUORENE (F) | 0.061 | 1.37 | 0.073 | 1.09 | 0.063 | L.D.L. |
| DIBENZOTHIOPHENE (DBT) | 0.034 | L.D.L. | 0.041 | L.D.L. | 0.036 | L.D.L. |
| PHENANTHRENE (P) | 0.088 | 13.08 | 0.093 | 10.14 | 0.095 | 15.21 |
| ANTHRACENE (Ant) | 0.032 | L.D.L. | 0.036 | L.D.L. | 0.032 | 5.42 |
| 1-METHYL PHENANTHRENE (1-MP) | 0.020 | L.D.L. | 0.023 | L.D.L. | 0.021 | L.D.L. |
| FLUORANTHENE (Fl) | 0.034 | L.D.L. | 0.042 | 5.78 | 0.037 | 45.82 |
| PYRENE (Pyr) | 0.024 | 5.00 | 0.029 | 7.53 | 0.027 | 39.12 |
| BENZO (A) ANTHRACENE (BaAnt) | 0.004 | 6.17 | 0.004 | 8.16 | 0.004 | 24.43 |
| CHRYSENE (Chr) | 0.004 | 2.52 | 0.005 | 3.09 | 0.005 | 12.32 |
| BENZO(b) FLUORANTHENE (BbFl) | 0.004 | 6.27 | 0.004 | 8.20 | 0.004 | 17.67 |
| BENZO(k) FLUORANTHENE (BkFl) | 0.004 | 3.70 | 0.004 | 4.38 | 0.004 | 11.24 |
| BENZO(e) PYRENE (BePyr) | 0.004 | 5.69 | 0.004 | 10.55 | 0.004 | 17.16 |
| BENZO(a) PYRENE /BaPyr) | 0.004 | 9.90 | 0.004 | 15.88 | 0.004 | 26.09 |
| PERYLENE (Per) | 0.004 | 5.58 | 0.004 | 9.01 | 0.004 | 8.20 |
| INDENO(1,2,3) PYRENE (Ipyr) | 0.003 | 7.91 | 0.003 | 8.70 | 0.003 | 19.17 |
| DIBENZO(a,h) ANTHRACENE (DBahAnt) | 0.004 | L.D.L. | 0.004 | 6.74 | 0.004 | 8.25 |
| BENZO(g,h,i) PERYLENE (BghiPer) | 0.003 | 6.94 | 0.003 | 8.30 | 0.003 | 16.15 |
| ΣPAHs | 0.461 | 74.12 | 0.525 | 107.6 | 0.467 | 266.2 |
D.L.= Detection Limit
L.D.L.=Low Detection Limit
SW=Seawater
S=Marine sediments
1= [ng mL-1]
2= [ng g-1 d.w.]
d.w.=dry weight
The results obtained from each individual PAH of marine sediments and surface seawater at each tested place are displayed in Table 1. It is clearly observed that in the surface water samples the predominant PAHs were compounds of low molecular weight (LMW) PAHs (2-3 rings). Specifically, largest values presented were for naphthalene (0.044 -0.063 ng mL-1), fluorene (0.061 - 0.073 ng mL-1), and phenanthrene (0.088 - 0.095 ng mL-1) while high molecular weight (HMW) PAHs (4-6 rings) were present at much lower concentrations (0.003 -0.005 ng mL-1). This suggests that the incomplete combustion of fossil hydrocarbons may be the predominant source of PAHs in surface water [20].
On the other hand, HMW-PAHs have prevalence in the sediment samples. Being the CFE site the one that contains the highest concentrations of PAHs, particularly, of fluoranthene (45.82 ng g-1), pyrene (39.12 ng g-1), and benzo (a)pyrene (26.09 ng g-1). It is known that LMW-PAHs are more easily biodegradable and consequently, predominant in seawater due to their partial solubility, whereas HMW-PAHs are recalcitrant and hydrophobic compounds, therefore, they can adsorb onto settling particles and reach the marine sediment bed [21,22].
Additionally, to evaluate potential petrogenic or pyrogenic PAHs sources in samples of sediments, FI/Pyr and BaAnt/Chr ratios were calculated. The Fl/Pyr index values obtained for CFE and C sampling sites were 1.17 and 0.76, respectively, while at M sampling site could not be calculated, since fluoranthene was below the detection limit. As for the BaAnt/Chr index, calculated values in three sampling sites were 1.96 (CFE), 2.64 (C) and 2.44 (M). Hydrocarbon source maybe pyrogenic, if ration is of Fl/Pyr >1 and BaAnt/Chr <2, but if values are <1 y >2, respectively, indicate a possible petrogenic source [23]. Indices values obtained in this work suggest that hydrocarbons present in CFE sampling site could be from pyrogenic source, while in M and C sampling sites, a petrogenic origin of these compounds is likely. This is in accordance with anthropogenic activities that are carried out in the coastal area of the Rosarito Port, where there are fuel discharges (M and C sampling sites) and boat traffic (CFE sampling site).
One of the most abundant HWM PAHs found in seawater and marine sediments was pyrene. Therefore, the isolated bacteria were considered as highly promising to degrade this pollutant. Despite the fact that pyrene per se is not carcinogenic compound [21,24], it has a chemical structure of four rings similar to several carcinogenic PAHs as benzo(a)pyrene which is highlights the importance to remove it of the environment [25]. Further, numerous studies on pyrene degradation by Mycobacterium spp. have been reported and their degradation pathways have subsequently been studied through chemical and omics analyses [23,26-28]. Biodegradation by bacteria has been further advanced through the documentation of new isolates that represent diverse bacterial types that have been isolated from different environments and that possess different metabolic capabilities [18,29,30]. Therefore, in this work we carried out the isolation of indigenous bacteria from seawater and marine sediments samples through the enrichment method using pyrene as the sole carbon and energy source.
Isolation and characterization of bacteria with high potential to degrade pyrene
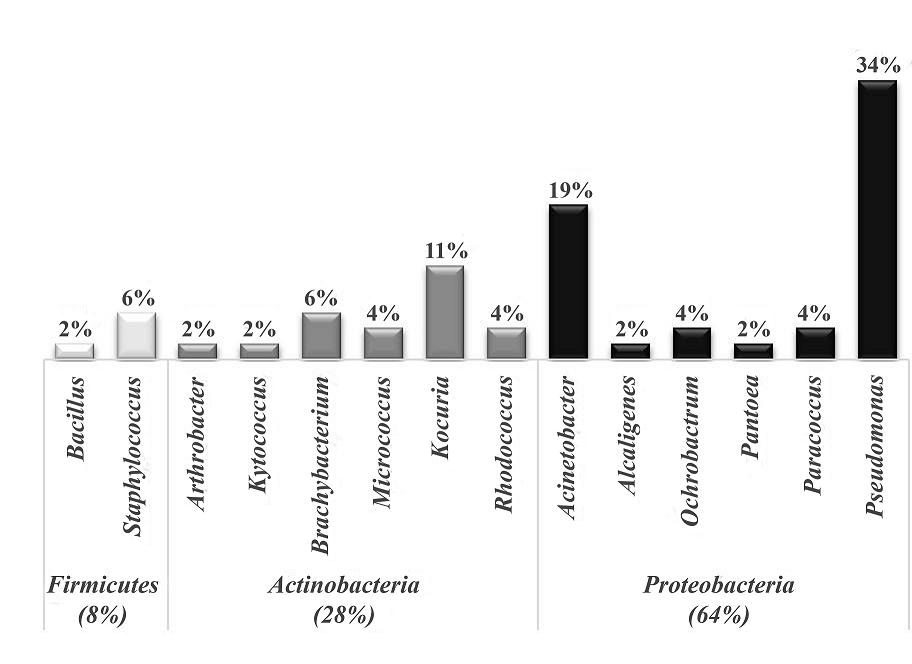
A total of 52 native isolates with capacity to growth in 25 mg L-1 of pyrene as sole source of carbon and energy were classified by MALDI Biotyper system. Initially, mass spectra obtained for each isolate were compared with reference database spectra (total of 5989 input). For this purpose, a pattern recognition algorithm uses peak positions, peak intensity distributions and peak frequencies, providing identification if the score value agree with the criteria established by the manufacturer [14]. Based on that, 12 of the isolates presenting the log(score) values between 1.70 and 1.99 were identified only at the genus level, whereas 19 of the isolates were assigned with the log(score) values ≥ 2.00 and ≤ 2.30, so the allocation was at the species level. Finally, 21 isolates presented a log(score) levels up to 2.30, with highly probable species identification (Supplementary Table S1). It is important to note that although the database used is mainly composed of organisms of clinical origin, score values with high probability of identification have been previously reported for environmental samples with results comparable equal or higher than those generated by 16S rRNA sequence [31,32].
The distribution of the population isolated of the culture enriched with pyrene is presented in Fig.1. The isolates fall into three large phyla: Firmicutes, Actinobacteria and Proteobacteria. The phylum Firmicutes was the least abundant with four isolates: one Bacillus sp. (2%) and three strains of Staphylococcus (6%). Curiously, despite the fact that Bacillus genus possesses a high metabolic versatility toward pyrene and other toxic compounds, it was found as the least abundant. Otherwise, some members of the Staphylococcus genus are well known human pathogens and also some of them as S. warneri have been reported to have the ability to degrade PAH [33]. In our case, for their ability to degrade PAHs [23,34,35]. and several reports proved utility of MALDI-TOFMS as a robust technique for identification of bacteria isolated from different environments [15,32,36,37]. Of note, is important to use the database of clinical origin for the identification of isolated microorganisms with potential use in bioremediation and at the same time discard those that may represent a risk to health.
Chemotaxonomy of isolates identified as Acinetobacter, Kocuria and Pseudomonas
Because Acinetobacter, Kocuria and Pseudomonas were the most abundant bacterial communities and in some isolate of Kocuria and Pseudomonas the log (score) was less than 2.0, a detailed spectra analysis of the protein profile was also performed using chemometric tools as PCA and CCI. The aim of this analysis was to inquire the feasibility of differentiation or typification of strains from the same species.
Typing of isolates identified the genus Acinetobacter, Kocuria and Pseudomonas
Acinetobacter. The genus Acinetobacter consists of non-fermenting coccobacilli, strictly aerobic, non-motile that are widely distributed in nature [38] and recently some strains obtained of soils have demonstrated a hydrocarbon-degrading capability [39]. Furthermore, several MALDI-TOF MS studies showed its ability to differentiate various Acinetobacter spp. as well as organisms that fall within the Acb (A. calcoaceticus- A. baumannii) complex those are closely from each other and are considered pathogenic species [40,41]. In our case, all the nine isolates identified as Acinetobacter showed a log (score) value ≥ 2 (Table S1) and present the phenotypic characteristics of the genus.
Figure 2a shows typical protein fingerprints of the region between 2,500 and 12,500 m/z obtained by MALDI-TOF MS of these nine isolates. The number of peaks in this range was similar for each of the strains, oscillating from 25-50. As an example, the peak at m/z 9481 was present in all the isolates, while the peaks at m/z 9114 and 9350, were only found in the isolated 1.37P; and peaks of m/z 10340 and 10472 were characteristic for the isolated 2.34P and 1.5P, respectively (Fig. 2a). To increase the allowance of peaks distinguishing and reducing the weighting of common peak in data sets from closely related organisms, a PCA-based approach was used (Fig. 2b). In the obtained model, the first three components accounted for 75% of the total data variability. The distribution of samples agreed to the species and three perfectly defined clusters are observed according with three different species: A. lwoffii, A. johnsonii and A. schindleri. Cluster analysis of PCA separated the 1.44P isolate identified as A. schindleri of the other two identified species. All isolates identified as A. johnsonii (1.12P, 1.15P and 1.42P) were classified in the same cluster located on the right of the first component. Whereas the 1.37P isolate was grouped distant from the rest of the isolates identified as A. lwoffii (1.5P, 1.6P, 1.43P, 2.34P). In summary, these results clearly suggest that the 1.37P isolate is a different strain of A. lwoffi and confirm the relevancy of Biotyper system for easily distinguish between species and biotyping of Acinetobacter strains.
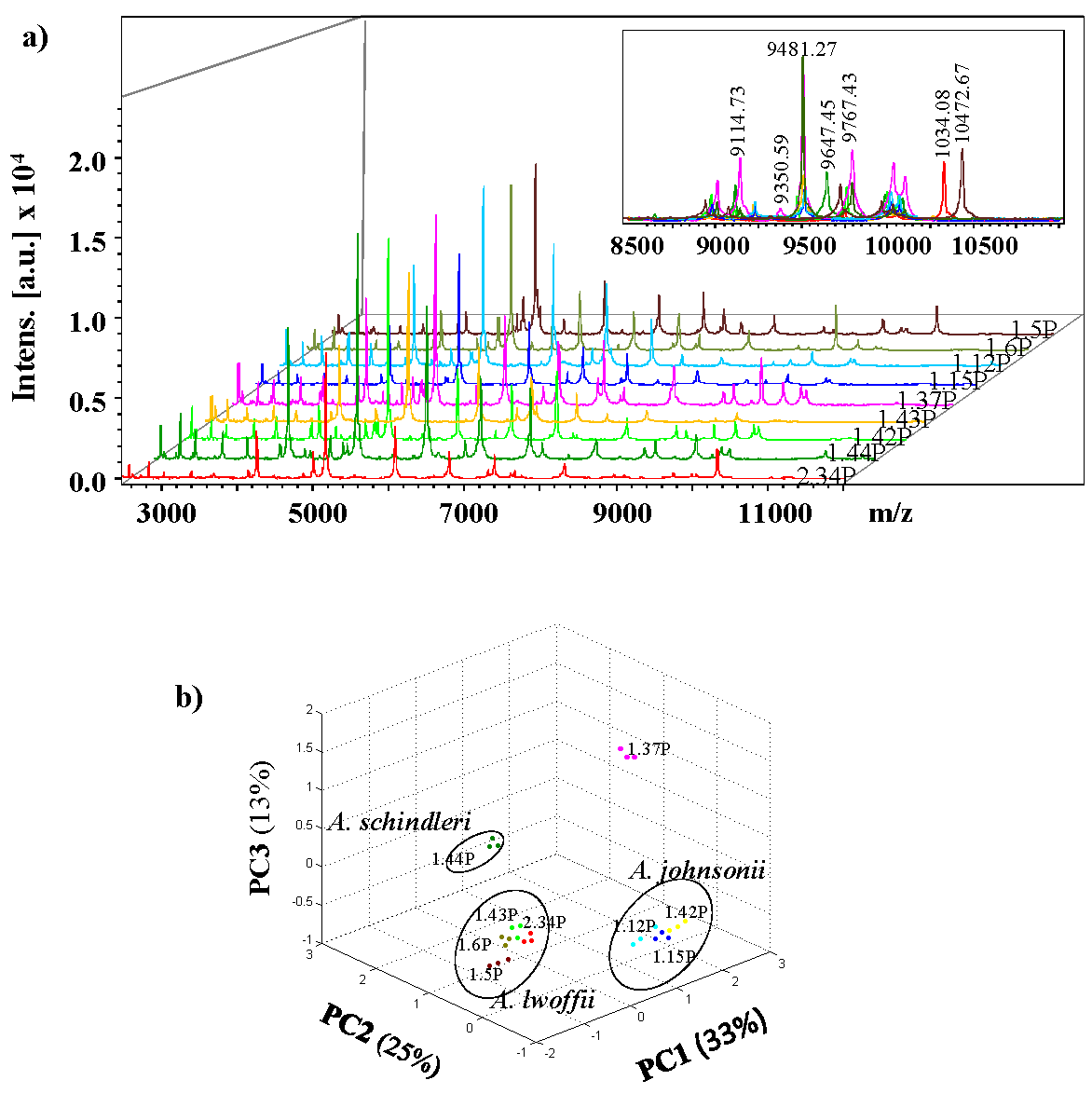
Fig. 2 Representative MALDI-TOF mass spectra of nine isolates identified as Acinetobacter and PCA 3D dot plots. a) Typical mass spectra of isolates 1.5P, 1.6P, 1.12P 1.15P, 1.37P, 1.42P, 1.43P, 1.44P, and 2.34P are presented in the m/z range of 2,500 to 1,2500 Da. b) PCA 3D dot plot of mass spectra obtained for the isolates of Acinetobacter using Biotyper software (version 4.0.14). The PCA was realized considering three biological replicas for each isolate.
Kocuria. This genus was created in 1995 after a detailed phylogenetic and chemotaxonomic analysis of the genus Micrococcus from which it was separated. Currently it contains 19 species with validly published names and classified into four clades based in 16S rRNA gene sequence [42]. The members of the Kocuria genus were rarely associated with human infection so they are excellent candidates for bioremediation processes. In fact, several strains of K. rosea isolated from soil can degrade LMW-PAHs [43,44].
The initial analysis of classification by MALDI-TOF mass spectrometry based on protein fingerprints using the Bruker database (22 reference spectra for different Kocuria strain) showed that isolates 1.28P, 1.32P, 1.33P, 1.36P, 2.10P and 2.18P, belong to the Kocuria genus (Table S1). Figure 3a shows mass spectra acquired for these strains in the m/z region 2,500 - 12,500. Macroscopic inspection of reveals both, common and differential peaks appearing in spectra of different strains. For example, m/z signals at 4687, 5126 and 5183 were found in the isolates identified as K. rosae whereas the m/z 5078 and 5348 characteristic for the isolated of K. rhizophila. Despite being classified as different species, the 1.32P isolate presents some common ions with 1.33P isolate such as m/z 4759, as well as unique peaks (m/z 5040).
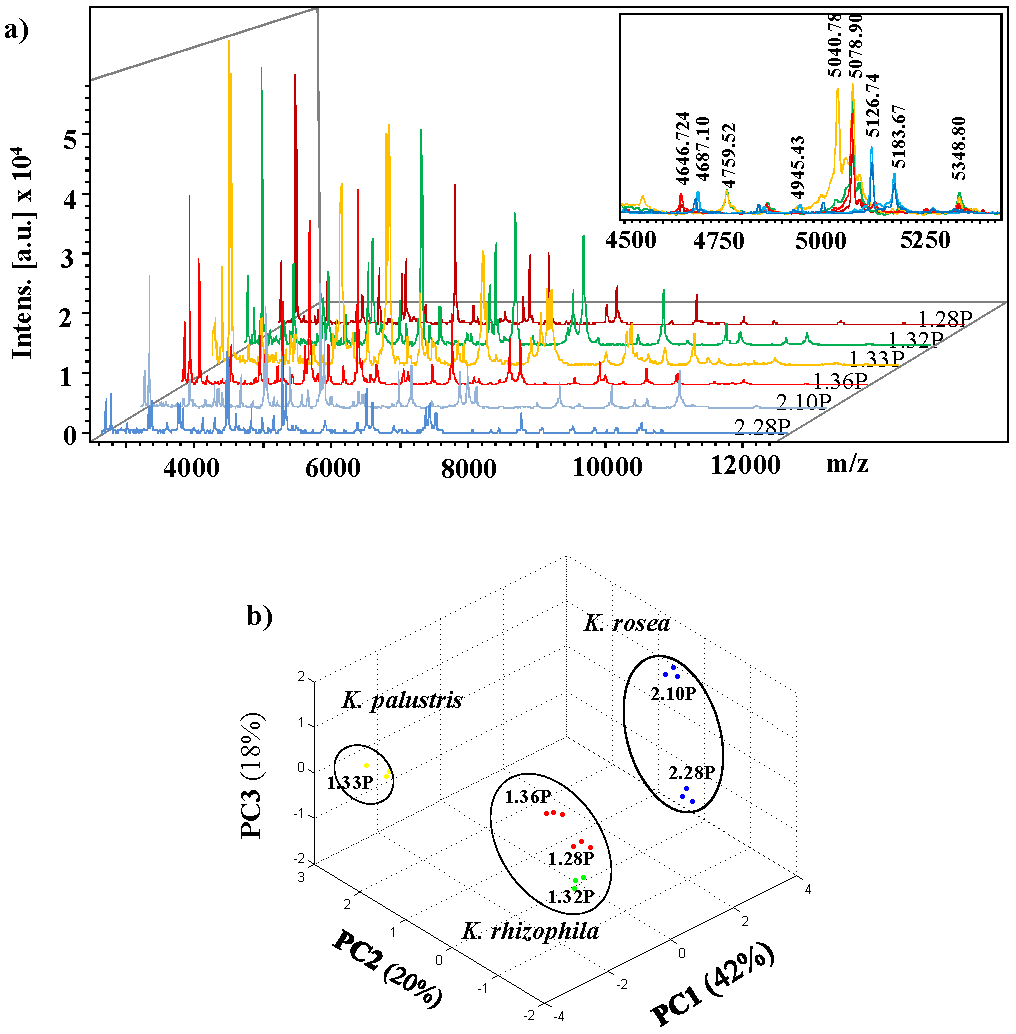
Fig. 3 Representative MALDI-TOF mass spectra of six isolates identified as Kocuria and PCA 3D dot plots. a) Typical mass spectra of isolate 1.28P, 1.32P, 1.33P, 1.36P, 2.10P and 2.28P are presented in the m/z range 2,500 - 12,500. b) PCA 3D dot plot of mass spectra of isolates of Kocuria using Biotyper software (version 4.0.14). The PCA was realized considering three biological replicas for each isolate.
Moreover, the PCA analysis of the MALDI mass spectrum was performed to determine the minimized differential peaks set and to investigate the relationships between the different strains of isolated that belong to Kocuria genus. The first three principal components contribute for 82% of the total data variability. Through the initial of identification analysis, the 1.33P isolate is classified as best match as K. palustri (log score =1.86) and as a second-best option as K. rhizophila (log score =1.86) but in the PCA analysis is separated from the 1.28P and 1.32P isolates which corresponds to K. rizhophila with a cutoff value above of 2.0. Whereas the 1.36P isolate with a log(score) of 1.86, was grouped together with isolates 1.28P and 1.32P. Otherwise, the isolates 2.10P and 2.28P are in the same cluster, suggesting that both isolates belong to K. rosea, although the 2.28P isolate has a lower cut-off value that recommended (log score=1.77). Together, these results suggest with high probability species identification of Kocuria by analysis chemotaxonomy.
Pseudomonas. Pseudomonas and Pseudomonas-like bacteria are a genus per excellence in biodegradation processes of various organic compounds, including PAHs, while many produce bioactive substances, enzymes, and biosurfactants, other Pseudomonas isolates are used for biological control of plant diseases and bioremediation [45,46] . Depending on the species of Pseudomonas MALDI-TOF MS performed equally as well as conventional systems for the correct identification the distinction at the level of species, subspecies, genomovars or strains. MALDI-TOF has gained popularity, and it is also a useful tool for the phenotypic characterization and identification of Pseudomonas strains, allowing the detection of Pseudomonas in environmental samples [32,47].
Similar as for typing Acinetobacter and Kocuria, we applied PCA of mass spectra obtained from 17 isolates identified as Pseudomonas like. In our analysis, most of the isolated were clustered into three groups defined as P. putida, P. aeruginosa and P. stutzeri (Fig. 4). The first three principal components described 82% of the total data variability.

Fig. 4 Principal components analysis (PCA) from mass spectra obtained by MALDI-TOF of 17 isolates identified as Pseudomonas using Biotyper software (version 4.0.14). The PCA was realized considering three biological replicas for each isolated.
The cluster analysis locates the 1.22P, 1.23P and 1.24P isolated in the P. putida group, however, the 1.22P isolate had previously been classified as P. mendocina with a log (score) of 2.22 (Table S1), so, its identification at the species level must be taken with reservation. Otherwise, the isolate 1.3P (P. oryzihabitans) is distributed in a space away from the rest of the isolates of P. putida group. Moreover, the 2.20P, 2.21P and 2.30P isolates were grouped within P. aeruginosa group as expected (Fig. 4).
Species of P. stutzeri and P. xanthomarina belong to P. stutzeri cluster. However, isolates 1.19P, 2.17P, and 2.18P that were previously identified as probable P. xanhthomarina with a log (score) ≤ 2.0(Table S1) were clearly separated: the 1.19P and 2.18P isolated are located between the positive side of PC2 and negative side of PC3, while 2.17P is located more positive side PC1 and less negative side of PC3, which suggests that these is probably a different strain of P. xanthomarina. The figure 4a shows that all isolates identified as P. stutzeri with a log(score) ≥2.25, are grouped in the same space. Moreover, according to the identification using the database, some of them it made a match with two different strains of P. stutzeri (Table S1).
Further study with mass spectra of the seven isolates as P. stutzeri, in order to check the relatedness between each set and discriminate between strains was conducted by CCI analysis available with MALDI Biotyper software (Table 2). CCIs for 2.3, 2.8, and 2.14 was a high match value (0.9) suggesting that it may be the same strain. The 1.9P presents no relationship (value 0.03) with the isolates 1.34P and 2.4P.
Table 2 CCI matrix value for the MALDI TOF MS data of P.stutzeri isolate.
| Isolate | 1.9P | 1.34P | 2.3P | 2.4P | 2.5P | 2.8P | 2.14P |
|---|---|---|---|---|---|---|---|
| 1.9P | 1.00 | ||||||
| 1.34P | 0.03 | 1.00 | |||||
| 2.3P | 0.65 | 0.70 | 1.00 | ||||
| 2.4P | 0.03 | 0. 72 | 0.51 | 1.00 | |||
| 2.5P | 0.74 | 0.73 | 0.59 | 0.80 | 1.00 | ||
| 2.8P | 0.71 | 0.75 | 0.96 | 0.59 | 0.68 | 1.00 | |
| 2.14P | 0.75 | 0.78 | 0.92 | 0.58 | 0.68 | 0.96 | 1.00 |
The CCI matrix was calculated with MALDI Biotyper 4.0.14 software with the default settings: the lower bound is 3,000-12500 Da, the resolution of the mass range is four, and the number of intervals for CCI is five. A CCI value near 1.0 indicates relatedness between the spectral sets, and 0 indicates no match.
In summary, in this work we found diverse genera of three large phylogenetic groups, which are ubiquitous in saline environments or in hydrocarbon-impacted sites. In addition, using a library of clinical origin, it allowed detect possible strains that can be a danger to health in a fast and reliable way.
Conclusions and Perspectives
Anthropogenic activities have contributed to the PAHs inputs in the coastal area of Rosarito Port, B.C., Mexico. A promising alternative to remove PAHs from this environment is by using autochthonous microbiota. However, it is important to identify microorganisms with potential use in bioremediation and, at the same time, discard those that may represent a risk to health. Biotyping by MALDI-TOFMS has provided compelling evidence of strain diversity coming from these environments with potential use in bioremediation in a way of low costs, fast and reliable. Additionally, the applying of chemotaxonomic techniques can detect very minor differences in the expression of major protein, allowing the characterization in some cases, at strain level. The above demonstrate that MALDI-Biotyper system is becoming an essential analytical tool in the environmental microbiology field and it can be considered an alternative for conventional biochemical and molecular identification in a microbiological laboratory. It should be noted that, as far as we know, it is the first report that generates information on cultivable bacterial diversity in areas impacted with PAHs in the coastal zone of Rosarito Port and in the Baja California Coast.
Finally, an excellent candidate to carry out bioremediation studies in saline environments is the Kocuria genus, because, despite members of this genus mostly have been reported as LMW-PAHs-degrading microorganisms, in this work, we have been able to obtain pyrene-utilizing Kocuria isolates and, in addition, they are not human pathogens, so, they could be applied in HMW-PAHs bioremediation.











 nueva página del texto (beta)
nueva página del texto (beta)