Introduction
Transition metal compounds play crucial roles as cofactors in metalloproteins, they act mainly as a Lewis acid. Two essential metal ions, namely zinc and copper ions, modulate enzymes activities, catalytic and regulatory functions, oxidative-reductive processes, etc. [1]. Zn(II) has a specific role in bioinorganic processes because of the peculiar properties of the coordination compounds of the zinc(II) ion, generally can be four-, five-, or six-coordinate, without a marked preference for six coordination [1,2]. As a catalytic cofactor, Cu(II) is required in metalloproteins and influences biological oxidation-reduction reactions and electron transfers thanks to the couple Cu(II)/Cu(I) [1].
Zinc is a good Lewis acid, especially in complexes with lower coordination numbers; it lowers the pKa of coordinated water and is kinetically labile, and the inter conversion among its four-, five-, and six-coordinate states is fast [3]. The theoretical studies have shown that zinc does not have a strong preference for a particular number of water molecules in its first coordination layer and can accommodate four, five, or six water ligands; the calculated energy differences between isomeric [Zn(H2O)6]2+, {[Zn(H2O)5]‚(H2O)1}2+, and {[Zn(H2O)4]‚(H2O)2}2+ complexes differ by only a few kilocalories per mole [4]. Moreover, dynamic conversion of structural zinc into a transient catalytic centre may be a mechanism for nucleic acid cleavage [5].
On the other hand, copper(II) controls cancer development. It serves as a limiting factor for multiple aspects of tumour progression, growth, angiogenesis and metastasis [6-8]. Recently, researchers have developed new complexes containing two copper(II) centres, which target two neighbouring phosphates oxygen on the DNA backbone that provide active sites for metalloenzymes such as nucleases [8]. The chemistry of copper is dominated by the +2 oxidation state, e.g. copper(II) complex ions. In comparison to other divalent first-row transition-metal aqua ions, the [Cu(H2O)6]2+ ion is extremely labile [9-12]. This effect is a consequence of Jahn-Teller distortion. As a result of the d9 electronic configuration, an elongation of the axial-bound solvent molecules are weakly coordinated. Due to this distortion the axial water molecules are weaker bound to the central atom and therefore can be more easily substituted [13]. The strong ligand field forces the metal ion into a different geometry, for example, 2,2′,2′′-triaminotriethylamine ligands (tren) will restrict the degree and rapidity of distortion of the [Cu(tren)H2O]2+ complex and remove the dynamic Jahn-Teller effect as labilizing effect [14]. The bulk of five-coordinate {Cu(terpy)(bipy)} and {Cu(terpy)(phen)} (terpy= 2,2′:6′,2′′- terpyridine or derivative, bipy= 2,2′-bipyridine or derivative, phen= 1,10-phenanthroline or derivative) complexes exhibiting ostensibly square-based pyramidal geometries also shows an additional interaction in the remaining axial site leading to a better description as their being six-coordinate [15].
The main goal of these studies was to investigate the kinetics and mechanism of ligand-substitution reactions between [ZnCl2(terpy)] and [CuCl2(terpy)] and biomolecules. In order to verify the changes in the coordination sphere that may occur on the side exposed to solvent, the structures of complexes were optimized by DFT method. As part of our interest of potential medical application of Zn(II) and Cu(II) complexes with chelating ligands, the antibacterial activity of these complexes have been investigated. It was envisaged that these studies could provide more information for understanding structure-reactivity correlation between [ZnCl2(terpy)] and [CuCl2(terpy)] complexes and biological relevant nucleophiles and could help us for the future design of novel zinc(II)- and copper(II)-based therapeutic drugs.
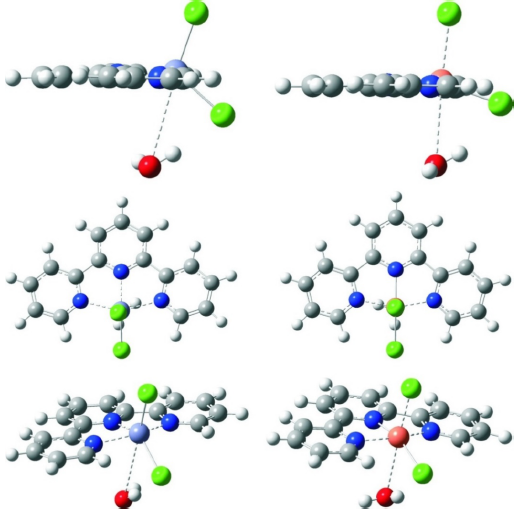
The ligand-substitution reactions were studied in aqueous solution at pH 7.38 as a function of nucleophile concentration at 22ºC by Uv-Vis spectrophotometric techniques. The structures of the complex and the selected nucleophiles are shown in Fig. 1.
Results and Discussion
Kinetics Studies
The reactions of five-coordinate [ZnCl2(terpy)] and [CuCl2(terpy)] complexes with bioligands can be monitored in the range of ca. 260-340 nm. An example of the Uv-Vis spectra and time trace are shown in Fig. 2. Solutions were prepared by dissolving known amounts of complexes in 0.005 mol L−1 phosphate buffer in the presence of 0.010 mol L−1 NaCl in order to prevent spontaneous hydrolysis reactions (see Supplementary Material, Figs. S1 and S2).

Fig. 2 Uv-Vis spectra recorded for the reaction of [CuCl2(terpy)] complex with guanosine-5’-monophospate at pH 7.38 (0.005 mol L−1 phosphate buffer) in addition of 0.010 mol L−1 NaCl at 22ºC: (a) spectra before the reaction; (b) spectra obtained after mixing of the reactants. Intercept: time trace obtained for the reaction at 330 nm.
All kinetic studies were performed at physiological pH, the presence of 0.010 mol L−1 NaCl. At higher concentration of chloride ( > 0.020 mol L−1 NaCl) an increase in absorbance was observed for the [ZnCl2(terpy)] complex (see Supplementary Material, Fig. S1). This may indicate the additional coordination of chloride ions in the solution, which could cause expansion of coordination sphere to six-coordinate. A six-coordinate complex can experience ligand dissociation, giving rise to a five-coordinate complex with little energy loss and then little energetic barrier [2]. The increase in absorbance for the [CuCl2(terpy)] complex have not been observed in the presence of higher chloride concentration (see Supplementary Material, Fig. S2).
The differences between spectra of investigated [ZnCl2(terpy)] or [CuCl2(terpy)] complex without chlorides and spectra of the solutions with various concentration of chlorides only can be noticed in intensity of the absorbance. The maximum positions have not been shifted due to coordination of the chloride, also the presence of the isosbestic point, which indicates the presence of different complex species and physical changes, has not been established (see Supplementary Material, Fig. S1 and S2). Thus, based on the analysis of experimental data, the existence of mixed Cl-H2O complexes was not observed.
The all kinetic traces gave excellent fits to a double exponential function. The so-obtained pseudo-first order rate constants, k obsd1 and k obsd2, calculated from the kinetic traces (absorbance/time traces) were plotted versus the concentrations of the entering nucleophiles. A linear dependence on the nucleophile concentration was observed for the reactions of [ZnCl2(terpy)] and [CuCl2(terpy)] complexes with DNA constituent (5’-IMP and 5’-GMP) and amino acids (L-Met and DL-Asp) (see Figs. 3 and 4).

Fig. 3 Pseudo-first order rate constants as a function of nucleophile concentration for the first reactions of the [ZnCl2(terpy)] complex with L-Met, 5’-IMP, 5’-GMP and DL-Asp at pH 7.38 (0.005 mol L−1 phosphate buffer) in the addition of 0.010 mol L−1 NaCl at 22ºC.
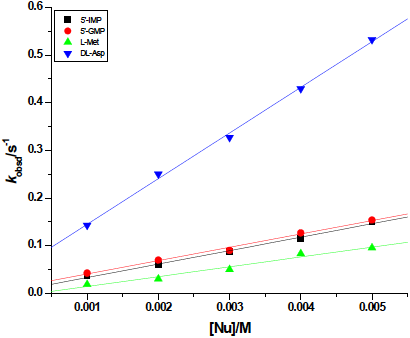
Fig. 4 Pseudo-first order rate constants as a function of nucleophile concentration for the first reactions of the [CuCl2(terpy)] complex with L-Met, 5’-IMP, 5’-GMP and DL-Asp at pH 7.38 (0.005 mol L−1 phosphate buffer) in the addition of 0.010 mol L−1 NaCl at 22ºC.
Observed pseudo-first-order rate constants, k obsd1 and k obsd2, depend on the entering nucleophile (Nu) concentration as given in Equation (1) and (2).
Linear fits passing through the origin for some reactions in present study indicating that possible parallel or backward reactions are insignificant or absent, i.e. k -1 and k -2 are negligible and Equations (1) and (2) simplify to k obsd1 = k 1[Nu] and k obsd2 = k 2[Nu]. Thus, in the present systems, direct nucleophilic substitution is the major observed reaction pathway under the selected conditions. The observed small intercepts for the dichlorido complexes are ascribed to the back reaction with the excess chloride present in solution. The derived rate constants are summarized in Table 1 and 2.
Table 1 Second-order rate constants and activation parameters for the reactions of the [ZnCl2(terpy)] complex with biomolecules: 5’-IMP, 5’-GMP, L-Met and DL-Asp at pH 7.38 (0.005 mol L−1 phosphate buffer) in the addition of 0.010 mol L−1 NaCl at 22°C

Table 2 Second-order rate constants and activation parameters for the reactions of the [CuCl2(terpy)] complex with biomolecules: 5’-IMP, 5’-GMP, L-Met and DL-Asp at pH 7.38 (0.005 mol L−1 phosphate buffer) in the addition of 0.010 mol L−1 NaCl at 22°C

Similar reaction pathways were obtained for the ligand substitution reactions between copper(II) complex with a sterically constrained pytBuN3 chelate ligand (pytBuN3 = 2,6-bis(3,5-di-tert-butylphenyliminomethyl)pyridine) and thiourea (TU) and N,N,N,N’-tetramethylthiourea (TMTU) [14a].
Substitution processes with DNA constituents and amino acids
The square pyramidal structure of Zn(II) and Cu(II) in biological systems prefers O-carboxylate, carbonyl and N-imidazole donor bioligand [1]. Inosine-5’-monophosphate (5’-IMP) and guanosine-5’-monophosphate (5’-GMP) can coordinate to metal ions via N1 and N7 [16]. Under our experimental conditions (pH 7.38) N7 atom is deprotonated [16]. It is also expected that at this pH the 5’-monophosphate residue of the nucleotide (pK a ≈ 6) is partially deprotonated. Binding through the N7 position of 5’-GMP in a neutral or weakly acidic medium to Zn(II) has been verified by 1H and 31P NMR spectroscopy [17]. The possibility of interaction of Cu(II) complex with oxygen from phosphate residue in DNA has been verified during the investigation of hydrolytic cleavage ability of transition metal ions, mainly because of the steric hindrance [18].
From a comparison of reactivity of the used complexes, it could be concluded that [CuCl2(terpy)] complex is at least 2-10 time more reactive than [ZnCl2(terpy)] (see Tables 1 and 2). The values of the second-order rate constants for the first reaction step of both complexes with 5’-IMP and 5’-GMP are equal order of magnitude (Table 1 and 2). The occurred similar reactivity of 5’-IMP towards [CuCl2(terpy)] for both reaction steps could be due to decreasing in the electronic density on the copper(II) center caused by π-acceptor ability of the tridentate chelate, 2,2′:6′,2”-terpyridine what caused that both chloride are equal for parallel substitution routes. The versatility of coordination in comparison to cisplatin-based drugs may provide access to the treatment of different cancer types with different toxicity [17]. According to the literature data, there are not many studies of the ligand-substitution reactions between Zn(II) and Cu(II) complexes and biologically relevant nucleophiles, thus the reactivity of bioligands was compared with obtained kinetic data for substitution reactions of [PtCl(terpy)]+ complex [17b)]. From comparison, it was observed that the order of reactivity of biomolecules in the substitution reactions with Pt(II) is the same and platinum(II) complex react slightly faster with DNA constituent.
Five-coordinate metal centers Zn(II) and Cu(II) according hard-soft acid nature of metals prefer O-carboxylate bioligands [1,16,19-23]. The coordination of L-Met and DL-Asp takes place via O-carboxylate donor atoms, the formation of chelate O-N-ammine haven’t been observed [21-23]. It is well known that coordination of zinc(II) ion to an L-aspartic acid through oxygen (O-Asp81) is present in Cu/Zn-superoxide dismutase (SOD1) [16,19]. Similar order of reactivity have been obtained for the first step between complexes and DL-Asp (for [ZnCl2(terpy)] k 1 = 79 ( 2 mol−1 L s-1; for [CuCl2(terpy)] k 1 = 96 ( 3 mol−1 L s-1). The rate constants demonstrate that the second substitution step is slowed down almost 10 times due to substitution of the first chloride in Zn(II) complex by DL-Asp. From the data in Tables 1 and 2, it can be concluded that the substitution reactions between Zn(II) and Cu(II) complexes with L-methionine are the slowest. At pH 7.38 the reactions occur between the zwitterionic form of L-methionine and complexes (the values for the dissociation constants for L-Met are pK a1 = 2.65 and pK a2 = 9.08 [24]).
The similar kinetic behavior of [ZnCl2(terpy)] and [CuCl2(terpy)] complexes suggests that the rates of nucleophilic substitution reactions are controlled by strong π-acceptor ability of the tridentate chelate 2,2′:6′,2′′-terpyridine, as well as, by steric hindrance and by electronic properties of the first coordinated nucleophiles. The [Zn(H2O)6]2+ and [Cu(H2O)6]2+ are well known for their extreme liability (k 298 > 107 s-1) with close to diffusion controlled rate constants [1]. The presence of bi-, tri-, tetra-dentate ligands in inner coordination sphere in complexes produce a decrease of the water exchange rates [1 , 14b, 14c]. The strong ligand field forces the metal ion into a different geometry. Furthermore, the rate constants of ligand-substitution reactions are decreased by a factor 103 because of this [14]. Based on above mention literature data, it can be concluded that square pyramidal coordination geometry around Cu(II) has higher impact on the rates of ligand-substitution reactions of [CuCl2(terpy)] complex than electronic properties and steric hindrance induced by the first coordinated biomolecules. This could be the reason why we obtained similar order of magnitude for the rate constants for both reaction steps with 5’-IMP and DL-Asp (see Table 2).
The activation parameters ΔH ≠ and ΔS ≠ were calculated by using an Eyring equation for the reactions with L-methionine at pH 7.38 (see, Supplementary Material, Fig. S5). All available activation parameters support an associative mechanism A or Ia for the both reaction steps (Tables 1 and 2). The significantly negative activation entropies suggest that the activation process in the studied systems seems to be strongly dominated by bond making [20a]. Thus, the transition states are expected to have six-coordinate character. The associative mechanism (A) for the substitution reaction processes is two-step pathway, involve formation of an intermediate with higher coordination number, followed by displacement of a weakly bound ligand. In many associative interchange reactions mechanism (Ia) well-defined intermediate is not observed and bond making between central metal ion and entering nucleophile dominate.
It is well known, if an associative mechanism is proposed, parallel pathways involving water and nucleophile attacks could be observed. In selected experimental conditions the presence of 0.010 mol L−1 NaCl was enough to suppress parallel hydrolytic pathway (see Supplementary Material, Figs. S1 and S2), direct nucleophilic attacks are major and take place in a manner as mentioned in Scheme 1. The mechanistic study of complex-ligand substitution reactions between five-coordinate [CuCl2(pytBuN3)] complex and TU, TMTU in the presence of 0.002 mol L−1 LiCl, showed an associative interchange (Ia) mechanism for the both chloride displacement [14a]. Similar results were reported for water exchange and ligand-substitution reactions on the five-coordinate [Cu(tren)H2O]2+ and substituted tren complexes [14b]. In this case, however, only one ligand can be replaced.

Scheme 1 The proposed reaction pathways for the reaction of the dichlorido Zn(II) and Cu(II) complexes with biologically relevant nucleophiles.
According to the literature data, the negative entropies of activation for displacement of both zinc ions from Zn2(ZP1) are consistent with an associative mechanism, in which the entering nucleophile attacks ZP1-bound metal (ZP1 = zinc fluorescent sensor of the Zinc pyr family). In this study nucleophiles tris(2-pyridylmethyl)-amine (TPA) and N,N,N′,N′-tetrakis(2-pyridylmethyl)ethylenediamine (TPEN), were used as alternative chelators for mobile zinc [20b]. The theoretical calculations on the water-exchange pathway for [Zn(H2O)4L]2+ ·2H2O (L = sp-hybridized N-donor ligands and sp2-, sp3-hybridized O-donor ligands) provide clear evidence for an associative (A) reaction mechanism as a result of moving one water molecule from the second coordination sphere to a vacant coordination site on the divalent zinc ion to form a six-coordinate intermediate [Zn(H2O)5(L)]2+ ·H2O [20c,20d].
Substitution processes with glutathione
Kinetics and mechanism of the substitution processes of [ZnCl2(terpy)] and [CuCl2(terpy)] complexes with tripeptide GSH were investigated under pseudo-first-order conditions with respect to the complex concentration. Whatever system is considered, the absorbance always shows an exponential growth or downtrend versus time indicating the first-order kinetics with respect to the nucleophile (v= -d[Nu]/dt = k obsd[Nu]) [20a]. The kinetics traces showed two reaction steps but different reaction mechanism for substitution reactions between [ZnCl2(terpy)] complex and glutathione has been obtained (Fig. 5)

Fig. 5 Pseudo-first order rate constants as a function of complex concentration for the first and second substitution reactions of the [ZnCl2(terpy)] and [CuCl2(terpy)] complex with glutathione at pH 7.38 (0.005 mol L−1 phosphate buffer) in the addition of 0.010 mol L−1 NaCl at 22°C.
For the substitution reactions between [ZnCl2(terpy)] and glutathione, first-order linear dependence, k obsd1 on the complex concentration at low concentration was observed. At higher concentration, saturation kinetics was obtained. These could be explained by considering that the first step is very fast pre-equilibrium formation of intermediate (pseudo-octahedral complex), followed by rearrangement to final complex whereas one chloride is substituted by GSH (Scheme 1). The value of rate second-order constant k 2 which described the substitution of the one chloride and pre-equilibrium constant K 1 could be determinate using Equation 3.
The obtained results are summarized in Table 3.
Table 3 Second-order rate constants for the reactions of the [ZnCl2(terpy)] and [CuCl2(terpy)] complexes with glutathione at pH 7.38 (0.005 mol L−1 phosphate buffer) in the addition of 0.010 mol L−1 NaCl at 22°C.
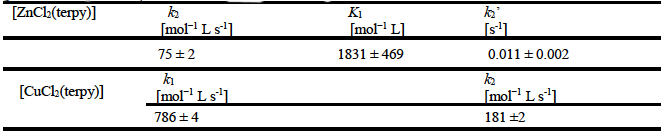
The value of pre-equilibrium constant was found to be K 1= 1831 mol−1 L (Table 3). The second substitution step is independent of glutathione concentration k obsd2= k 2’, which indicates that chelation process takes place (Scheme 1, Fig. 5). At pH 7.38 GSH is deprotonated [24], formation of five-membered chelate ring is possible via O-carboxylate and N-ammine group from (-glutamyl residue [25]. The rise and fall of the absorbance at 281 nm as a function of time is characteristic for a reaction that involves an intermediate in the overall process during which reactant is transformed into product (see Supplementary Material, Fig. S6).
For all the studied reactions between [CuCl2(terpy)] and glutathione an linear dependence on the concentration of complex was observed. Fits passing through the origin indicating that possible parallel reactions are insignificant or absent (see Fig. 5). The observed pseudo-first-order rate constants, k obsd1 and k obsd2, as a function of the total concentration of complex could be described by Equations (1) and (2), if we consider instead of concentration of Nu the concentration of [CuCl2(terpy)]. From the data in Table 3, it can be concluded that glutathione is the best nucleophile and Cu(II) complex has shown 10 times higher affinity to glutathione then Zn(II). Moreover, this result could be very important because copper and zinc balance is an important factor in maintaining glutathione levels. Glutathione may play a role in removing excess of copper and has little or no effect on Zn(II) transport inside cells [26]. In the cancer therapy glutathione has been used as a protecting agent and is administered before or after cisplatin [1]. At the same time platinum drugs could be inactivate in the reaction with glutathione, because Pt(II) complexes have high affinity to S-thiol group [1]. In order to explain the fact that Zn(II) and Cu(II) complexes react slower with glutathione then Pt(II), it is necessary to take into account the various factors, such as, the hard-soft nature of the metals, the versatility of coordination of the biomolecules and different geometrical structures of complexes [17b].
Antibacterial activity
Antibacterial activities of [ZnCl2(terpy)] and [CuCl2(terpy)] complexes were tested against seven strains of bacteria. The results of tested bacterias are presented in Table 4. Tetracycline antibiotic doxycycline was used as antibacterial control drug. For comparison, in the table the results of activities of this antibiotic also are given. The solvent (10% DMSO) had no effect on the growth of tested bacteria. Antibacterial activities of tested complexes were assessed by determining the MIC and MBC.
Table 4 Antibacterial activities of [ZnCl2(terpy)] and [CuCl2(terpy)] complexes against tested bacteria based on microdilution method.

aMinimal inhibitory concentration (MIC).
bMinimum bactericidal concentration (MBC) values are given as mg/mL for [ZnCl2(terpy)] and [CuCl2(terpy)] complexes and μg/mL for antibiotic (doxycycline).
The values of minimum inhibitory concentrations of complexes obtained in this experiment range from 1.25 mg mL-1 to >20 mg mL-1. According to the results from Table 4 the investigated complexes were more effective against Gram-positive than Gram-negative bacteria. The most active complex was [CuCl2(terpy)] complex (Table 4). The best effect was exhibited against Sarcina lutea (5 mg mL-1). Escherichia coli showed low sensitivity to both complexes. The study of the antibacterial activity suggests an absence of permeability of the complexes through the membrane proteins. Both complexes were more efficient against Escherichia coli ATCC 25922 in comparison with doxycycline.
Similar studies have shown that two new synthesized Cu(II) and Zn(II) complexes with xylitol have antibacterial effect against Pseudomonas aeruginosa and Candida albicans. Both copper and zinc complexes presented higher MIC against Pseudomonas aeruginosa than the free xylitol, showing that these complexes could be better local antibacterial compounds than xylitol [26]. The square pyramidal [Cu(lmx)(phen)(NO3)]·5H2O has shown minimum inhibitory concentration (MIC) in variousE. colistrains and comparison with free lomefloxacin indicated that the Cu(II) complex is an antimicrobial agent that is as efficient as the free antibiotic, but strongly suggested that the cell intake route of both species is different [27,28].
DFT Calculations
To clarify some doubts that we encountered during the experiment such as, is there possibility of coordination sphere expansion in [ZnCl2(terpy)] and [CuCl2(terpy)] complexes during investigation of the substitution processes with biomolecules, and in order to verify changes in the structure of investigated complexes in crystal and in solute forms, we optimized the structures using DFT method. The complexes have different central metal ions and have labile sites which can take part in ligand exchange interaction with biomolecule.
Comparing values of bond distances and angles of molecules optimized in vacuum with literature data of X-ray structural analysis, we confirmed very good agreement among theoretical and experimental obtained data (Table 5) [29, 30]. For both complexes the optimization was started from trigonal bipyramidal and from square pyramidal geometry as initial geometries. In both cases complexes have shown the same, following presented geometries. The way of atoms labeling in the investigated complexes is presented in Fig. S7, Supplementary Material.
Table 5 Some experimental and theoretical values of bond distances [Å], angles [⁰, labeled with A in front of brackets], and dihedral angles [⁰, labeled with D in front of brackets] of investigated complexes optimized in gas phase, and of monohydrate complexes optimized in water. M indicates metal, and O indicates oxygen atom of water molecule
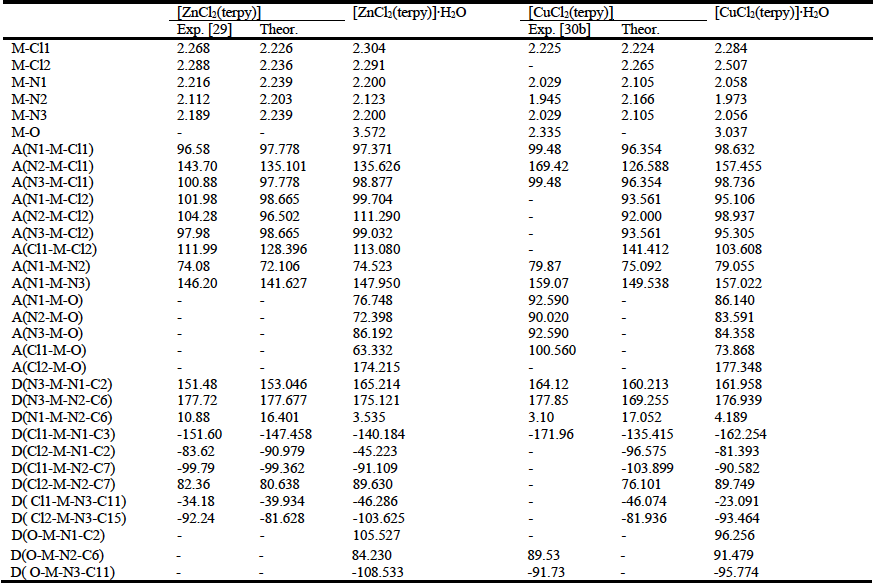
By comparing calculated bond distances, angles and dihedral angles with experimental obtained (Table 5), it was shown that optimized geometries give very good agreement with geometry identify by the X-ray crystallographic analysis [29]. Small discrepancies between theoretical and experimental results for complex are probably the result of dissimilarity in complex structure [30b]. Water molecule, as one of the ligands in experimental identified complex [30b)], has different steric and electronic properties than chloride atom. Therefore in this case the ideal agreement is neither to be expected.
Angle and dihedral angle values indicate that ligand terpyridine is not completely planar. Values of N2-M-Cl1 and N2-M-Cl2 angles, as of Cl1-M-N2-C7 and Cl2-M-N2-C7 dihedral angles indicate discrepancy of planarity (Table 5). Therefore, geometries of both complexes can be defined as a shaped square pyramidal (Fig. 6).

Fig. 6 Geometries of investigated complexes optimized in gas phase from three different perspectives: [ZnCl2(terpy)] - left and [CuCl2(terpy)] - right.
Approval that the optimized structures correspond to the real structures of molecules is obtained by comparing the calculated UV-Vis spectra with the experimentally obtained ones for aqueous solutions of molecules. UV-Vis spectral analysis was performed using Gaussian 09 [31]. Spectral analysis and visualization was performed using ChemCraft 1.6 [32a]. There is acceptable agreement between experimental and calculated spectra obtained for hydrated and non-hydrated complexes in aqueous solution (see Fig. S8, Tables S11 and S12, Supplementary Material). The intensities and positions of the peaks are in good agreement with experimental data. The most important charge transfers for both complexes regards to transitions of electron from lone pair of chloride atoms to metal.
It was impossible only on the basis of UV-Vis spectral analysis to make conclusion whether complexes during solvation bind water molecules as ligand or not. Those answers we tried to find from zero-point energies, enthalpy and Gibbs free energy of hydrated and non-hydrated molecules optimized in water as solvent. Based on the values of energy presented in Table 6, it can be seen that monohydrates are little stable than solvated non-hydrates for energy of water molecule. If we compare energies of monohydrates with sum of energies of solvated non-hydrate and water, it can be seen that enthalpies of monohydrates are lower for about 14 kJ mol-1. Values of Gibbs free energies for monohydrates are the consequent of entropic factor. Based on energetic stability of complexes, it can be concluded that both of complexes can make hydrates very easy, and that probably exist in dynamic equilibrium in aqueous solutions as anhydrated and monohydrated complexes.
Table 6 Absolute values of zero-point energy (ZPVE), enthalpy (H) and Gibbs free energy (G) for water molecule, and relative values of zero-point energy (ΔZPVE), enthalpy (ΔH) and Gibbs free energy (ΔG) for investigated complexes in aqueous solution

From data obtained using NBO analysis it can be seen that bonds between metal ion and ligands are mostly the result of electron transfer of lone pair electrons of nitrogen atoms, or unpaired electrons of chloride to the empty s orbital of metal ion (Tables S13, S14 and Fig. S9) [32b]. The energies of those bonds are higher than the energies of π - π* electron transfer. When water molecule enters into the coordinate sphere of complex, it builds very weak bond with metal ion, as a result of lone pair electron transfer from oxygen to the unoccupied orbital of metal. The low strength of this bond indicates easiness of hydratation and dehydratation of investigated complexes, as a result of dynamic equilibrium. From obtained results, could be concluded that the H-O-H…Cl non-covalent interactions are present in the solution for the both complexes.
Information about geometry of hydrated complexes, we obtained from values of angles and dihedral angles for monohydrate complexes presented in Table 6. Due to the entering of a water molecule into the coordinate sphere of the complex, the ligand terpyridine became more planar than in non-hydrated complexes. The central metal ion is little moved above this plane. N2-M-Cl2 angles deviate from the normal angle, as the N2-M-O angles. It was established that during formation of monohydrate, both complexes obtain little shaped octahedral geometry, with three nitrogen and one chloride atom in the central plane, and with water molecule and the other chloride atom on the line almost normal to the plane (Fig. 7).
Experimental
Chemicals
The nucleophiles inosine-5-monophosphate sodium salt hydrate (5’-IMP), guanosine-5-monophosphate sodium salt hydrate (5’-GMP), L-methionine (L-Met), DL-aspartic acid (DL-Asp) and glutathione (GSH) were obtained from Sigma-Aldrich, Acros Organics and Fluka. Nutrient liquid medium, a Mueller-Hinton broth was purchased from Liofilchem (Italy), while dimethyl sulfoxide (DMSO) was obtained from Sigma-Aldrich. An antibiotic, doxycycline, was purchased from Galenika A.D. (Belgrade, Serbia). Nucleophile stock solutions were prepared shortly before use by dissolving the chemicals in purified water. All other chemicals were of analytically reagent grade. Highly purified, deionized water was used in the preparation of all solutions. For the investigation the ligand substitution reactions at pH 7.38 a freshly prepared 0.005 M phosphate buffer was used. NaCl was used to adjust the chloride concentration.
Synthesis of complexes
The complexes [ZnCl2(terpy)] and [CuCl2(terpy)] were prepared according to the literature method [33,34]. Anal. N 11.21%, C 48.31%, H 3.12 %, calcd for C15H11Cl2N3Zn, N 11.37%, C 48.75%, H 3.00%. Found. Anal. N 16.97%, C 48.35%, H 3.11%, calcd for C15H11Cl2N3Cu, N 17.28%, C 48.99%, H 3.02%. The crystal structures of investigated complexes are square pyramidal with distortion, they are already known and published [29,30].
Instrumentation
Chemical analyses were performed on a Carlo Erba Elemental Analyser 1106. UV-Vis spectra were recorded on Uvikon XS, Hewlett-Packard 8452A diode-array and Shimadzu UV250 diode-array spectrophotometers in thermostated 1.00 cm quartz Suprasil cells. The temperature was controlled throughout all kinetic experiments to ± 0.1 °C. All kinetic measurements were performed under pseudo-first-order conditions, i.e., at least a 10-fold excess of the entering nucleophile or complex was used.
Kinetics measurements
Spectral changes resulting from mixing [ZnCl2(terpy)] and [CuCl2(terpy)] and nucleophile solutions were recorded over the wavelength range 200 to 800 nm to establish a suitable wavelength at which kinetic measurements could be performed (see Fig. 2, Supplementary Material, Table S1-S10). The ligand substitution reactions were studied for the nucleophiles: 5’-IMP, 5’-GMP, L-Met, DL-Asp and GSH. Reactions were initiated by mixing equal volumes of the complex and ligand thermostated solutions in the UV-Vis spectrophotometric cell and were followed for at least four half-lives. All kinetic experiments were performed under pseudo-first-order conditions with respect to the nucleophile concentration expect for the investigation the reaction between Cu(II) and Zn(II) with GSH where the concentration of the complex was at least a 10-fold excess. The measured k obsd values are summarized in Tables S1 - S10 (see Supplementary Material). All kinetic runs could be fitted to a double exponential function. The observed pseudo-first-order rate constants, k obsd, were calculated as the average value from three to four independent kinetic runs. The reactions were studied at pH 7.38 (0.005 mol L−1 phosphate buffer) at 22ºC in the presence of 0.10 mol L−1 chloride concentrations.
Suspension preparation for determination of antibacterial activity
Bacterial suspensions were preparated by the direct colony method. The turbidity of initial suspension was adjusted by comparing with 0.5 McFarland's standard [35], and they contain about 108 colony forming units (CFU)/mL. The 1 : 100 dilutions of initial suspension were additionally prepared into sterile 0.85% saline.
Microdilution method
Antimicrobial activity was tested by determining minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC). The stock concentration of the complexes was 20 mg mL-1. Two-fold serial dilutions of complexes were made in sterile 96-well plates containing Mueller-Hinton broth. The tested concentration range was from 10 mg mL-1 to 0.08 mg mL-1. The bacterial growth was monitored optically. The inoculated plates were incubated at 37ºC. The MIC concentration was defined as the lowest concentration of tested substance that visibly inhibited colonial growth. Minimum bactericidal concentration was determined by plating 10 mL of samples from wells, sample with no growth (no colony forming) was defined as minimum bactericidal concentration. Doxycycline dissolved in nutrient liquid medium, was used as positive controls. Stock solutions of crude extracts were obtained by dissolving in DMSO and then diluted into Mueller-Hinton broth to achieve a concentration of 10% DMSO. Solvent control test was performed to study the effects of 10% DMSO on the growth of bacteria. It was observed that 10% DMSO did not inhibit the growth of bacteria. Also, in the experiment, the concentration of DMSO was additionally decreased because of the two-fold serial dilution assay (the working concentration was 5% and lower). Each test included growth control and sterility control. All tests were performed in duplicate and MICs were constant.
DFT method
Optimization of complexes, as NBO and UV-Vis spectral analysis are done using M06 functional [36] implemented in Gaussian 09 program package in combination with 6-311++G(d,p) basis set [31] . Applied functional is tasted and confirmed by its developers as one of the best functional for a combination of main-group thermo-chemistry, kinetics and noncovalent interactions, as for the study of organometallic thermo-chemistry [36]. Simulation of the presence of water as the solvent is achieved by utilizing PCM/SMD solvation model. The absence of imaginary frequencies in optimized structures confirmed structure geometry with energy minima. NBO analyses of complexes is performed [32b].
Conclusions
The kinetics and mechanism of chloride substitution in [CuCl2(terpy)] and [ZnCl2(terpy)] complexes by biologically relevant ligands were studied in detail as a function of entering nucleophile concentration and temperature at pH 7.38. The kinetics showed that the substitution reactions involve the consecutive displacement of both chloride ligands. Higher reactivity of [CuCl2(terpy)] then [ZnCl2(terpy)] was obtained. The reactivity of the bioligands toward [CuCl2(terpy)] for the first reaction step follows the sequence GSH> > DL-Asp > 5’-GMP > 5’-IMP > L-Met, while toward [ZnCl2(terpy)] the order is DL-Asp > GSH > 5’-GMP > 5’-IMP > > L -Met. The π-acceptor properties of the tridentate N-donor chelate (terpy) predominantly control the overall reaction pattern. The different mechanism for complex-ligand substitution reactions between five-coordinate complexes and GSH has been obtained. The activation parameters ΔH ≠ and ΔS ≠ were calculated by using an Eyring equation for the reactions with L-methionine at pH 7.38 support an associative mechanism A or Ia for the both reaction steps. The stronger antibacterial activity of [CuCl2(terpy)] than [ZnCl2(terpy)] was obtained. This can be in correlation with a stronger affinity of Cu(II) for biomolecules. Absence of permeability of the proteins through cell membrane is probably due to the low thermodynamic stability of the prepared complexes and their dissociation in the solution. Based on energetic stability of complexes, it can be concluded that both of complexes make hydrates very easy, but the bond between water molecule and metal ion is pretty weak. Also, there is very good agreement between experimental and calculated spectra obtained for hydrated and non-hydrated complexes in aqueous solution. During formation of monohydrate, Zn(II) and Cu(II) complexes obtain little shaped octahedral geometry, with three nitrogen and chloride atom in the central plane, and with water molecule and the other chloride atom on the line almost normal to the plane.
Supplementary Material
Tables with pseudo-first-order rate constants (Tables S1 to S10); Tables with experimental and calculated electronic transitions (Tables S11 to S14). Figures with effect of chloride concentration on spontaneous hydrolysis of complexes (Figs. S1 and S2); Figures with pseudo-first order rate constants as a function of nucleophile concentration for the second reactions (Figs. S3 and S4); Figure with Eyring plots for the two reaction steps (Fig. S5); Figure with time traces obtained for the reaction of 0.02 mM GSH and 10 and 30-fold excess of the concentration of [ZnCl2(terpy)] complexes (Fig. S6); Atomic labeling of investigated complexes (Fig. S7); Experimental and calculated UV-Vis spectra of investigated complexes (Fig. S8); Changes of natural charge in ligands and central metal ions during complexation (Fig. S9).











 nueva página del texto (beta)
nueva página del texto (beta)