Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO
Share
Journal of the Mexican Chemical Society
Print version ISSN 1870-249X
J. Mex. Chem. Soc vol.56 n.1 Ciudad de México Jan./Mar. 2012
Article
Secondary Ligand Effects on the Cytotoxicity of Several Casiopeína's Group II Compounds
María Elena Bravo–Gómez,1 Silvia Dávila–Manzanilla,1 Jessica Flood–Garibay,1,2 Miguel Ángel Muciño–Hernández,1 Ángel Mendoza,3 Juan Carlos García Ramos,1 Rafael Moreno–Esparza,1 Lena Ruiz–Azuara*1
1 Departamento de Química Inorgánica y Nuclear, Facultad de Química, Universidad Nacional Autónoma de México, Avenida Universidad 3000, 04510, México D. F., México. Phone: +52(55)56223529; ruizazuara@gmail.com
2 Departamento de Ciencias Químico–Biológicas. Escuela de Ingeniería y Ciencias. Universidad de las Américas–Puebla, Ex hacienda Sta. Catarina Mártir S/N, 72810, Cholula, Puebla, Pue., México.
3 Centro de Química, ICUAP, Benemérita Universidad Autónoma de Puebla, 72570, Puebla, Pue., México.
Received May 19, 2011.
Accepted December 14, 2011.
Abstract
The aim of this study is to identify the influence of the ligand aminoacidato on the antiproliferative activity against human tumor cell lines HeLa, HCT–15 and SKL–U through the analysis of a set of Cu(II) coordination compounds with general formula [Cu(4,7–dimethyl–1,10–phenanthroline)(N–O)]NO3where N–O represents several essential aminoacids. Results show that the hydrophobicity of the amino acid ligands increases the cellular uptake of copper (II) in HeLa tumor cell line, however, the increase of copper concentration inside the cell is not enough to produce significant changes in antiproliferative activity in this tumor cell line. Thus, the substituents on diimine ligand apparently play the stronger role to control the activity of these complexes. Also molecular structures of compounds 2 (Cas IIala) and 4 (Cas IInva) are presented.
Key words: Copper, mixed chelate, aminoacids, ligand effect, inorganic anticancer compounds, cytotoxicity, intracellular distribution.
Resumen
El propósito de este estudio es el identificar la influencia del ligante aminoacidato en la actividad antiproliferativa a partir del análisis de una serie de compuestos con variaciones en la cadena lateral del aminoácido. Los resultados muestran que la hidrofobicidad del ligante incrementa la incorporación celular de Cu (II); sin embargo, no se ha encontrado una correlación entre la concentración intracelular de cobre con la actividad biológica. Los sustituyentes en los ligantes diimina cuentan con la mayor participación en el control de la actividad biológica. También se informan dos estructuras cristalinas de los compuestos 2 (Cas IIala) y 4 (Cas IInva).
Palabras clave: Cobre, chelate mezclado, aminoácidos, efecto ligante, componentes inorgánicos anticáncer, citotoxicidad, distribución intracelular.
Introduction
In the last decades metal complexes have gained a growing interest as pharmaceuticals for their use as diagnostic agents or as chemotherapeutic drugs [1–4]. In this field, one of the main goal is the development of non–platinum compounds with antineo–plastic activity through modes of action different from that of cisplatin (coordinated to DNA), for example, Sn(IV) [5], Ti(IV) [6], V [6], Au(III) [7], Ru(II) [8], Pd(II) [9], Rh(I) [10] and Cu(II) [11–13] compounds, involve non–traditional interaction with DNA or other biological targets as mitochondria. These new molecules have to fulfil several requirements in preclinical and clinical studies in their way to clinical phases, such as to reduce the toxicity; increase activity compared with known compounds, proved activity on cell lines resistant to known treatments and have preferentially low production costs.
Currently, several groups of copper complexes in both oxidation states (I) and (II) have been studied as potential antitumor agents [12–15]. Copper is an essential trace element important for the function of several enzymes involved in energy metabolism, respiration and DNA synthesis in the cell [16, 17], and the major functions of biological–active copper compounds involve redox reactions. The toxicity of copper comes from its ability to produce reactive oxygen species (ROS), displace other metal ions, peroxidize lipids and directly cleave DNA and RNA [18–23].
All these properties were taken into account in the design of copper (II) coordination compounds recorded and patented [24–26] under the name Casiopeínas® with general formulae [Cu(N–N)(N–O)]NO3 and [Cu(N–N)(O–O)]NO3; where the primary ligand N–N = non substituted and substituted 2,2'–bipyridine (bipy) or 1,10–phenanthroline (phen); and the secondary ligand N–O = α–aminoacidate or a peptide and O–O = acetyl–acetonate (acac) or salicylaldehydate.
Two compounds of this family [Cu(4,4'–dimethyl–2,2'–bipyridine)(acac)]NO3 (Casiopeína III–ia) and [Cu(4,7–dimethyl–1,10–phenanthroline)(glycinate)]NO3 (Casiopeína II–gly) have been evaluated in vitro and in vivo showing cytotoxic [27–29], genotoxic [27] and antineoplastic [30, 31] activity. The action mechanism is still not completely elucidated. However, there is evidence that supports the idea that these compounds are able to inhibit cell proliferation and produce cell dose–dependent death by apoptosis through mechanisms dependent and independent of caspase activation [31, 32]. The apoptosis observed might be the result of several processes like generation of reactive oxygen species (ROS) [27, 33, 34] or mitochondrial toxicity [31, 35, 36], which may act alone or in concomitance to produce cell death. In presence of reducing agents the cell growth inhibition [21, 24], and degradation of DNA [20, 21, 25] are potentiated, simultaneously with ROS increment, which suggest that DNA oxidation in cells may also be involved in cell death. Besides, it is possible that these compounds interact with DNA by intercalation due to the planar moiety corresponding to the diimine or the acetylacetonate ligand [34, 37–39].
In a QSAR study [40], the antiproliferative activity on HeLa, SiHa, HCT–15 and MCF–7 cells for compounds with general formula [Cu (N–N) (acac)] NO3 and [Cu(N–N)(glycinato)]NO3 was analyzed. According with QSAR studies the presence of the central fused aromatic ring in the phen containing complexes is necessary to preserve the antiproliferative activity. The IC50 has a strong relationship with the half–wave potential (E1/2) of the copper (II) center; being the most active those complexes which are weaker oxidants. A change in the secondary ligand from acac to glycinato has less influence on the biological activity when compared to the changes on the diimine ligand. It has been proposed that the role of the secondary ligand on antiproliferative activity is to assist the transport of the copper coordination compound across membranes due to an increment in hydrophobicity, acting as carriers in the uptake of copper (II). It is also possible that the modification of hydrophobic properties become more important by influencing directly the antiproliferative activity on a whole organism, where this property plays a major role in drug transport to the site of action. However, the exact contribution of the secondary ligand to the activity remained to be fully understood.
With the aim of understanding the role played by the secondary ligand on the biological activity shown by these coordination compounds, 13 copper (II) compounds with general formula [Cu(4,7–dimethyl–1,10–phenanthroline)(α–aminoacidato)]NO3 were synthesized, characterized and their antiproliferative activity was evaluated in three human tumour cell lines HeLa, HCT–15 and SKL–U. The structural variants in these complexes are only in the α–aminoacidato side–chain (Fig. 1).
Results and Discussion
For analysis purposes, complexes are classified in two groups: group 1, aliphatic aminoacids (compounds 1–8); and group 2, aminoacids with aromatic or heterocyclic rings (compounds 9–13). In the first group the hydrophobicity is enhanced with the increment of carbon chain length which allows analysing the effect of this property on the antiproliferative activity. In the second group, the structures are more varied and the electronic, steric and hydrophobic properties are modified simultaneously; however, they all share certain structural restrictions due to aromatic or heterocyclic rings. Results show that the changes performed on aminoacidate ligand have less influence on the activity than those performed on diimine. Complexes with aromatic or heterocyclic rings are more active than those with aliphatic aminoacids. The hydrophobicity enhancement due to the increase of carbons in the side–chain of aliphatic aminoacidates increases the cellular copper intake; however, the correlation with Log P or IC50 is not direct.
The complexes synthesized exhibit IR absorption bands typical for coordinated ligands, 4,7–dimethyl–1,10–phenanth–roline absorption bands are present in 1630, 1520, 1435, 885 and 725 cm–1. Aminoacidate ligands shown absorption bands around 3400–3200, 1600–1570 and 660–650 cm–1 for amines and 1640–1600, 1400–1380, 800–730, 565–540, 490–460 cm–1for caboxylate moiety. NO3– as counter ion (≈ 1384 cm1) and water molecules.
Compounds with aliphatic aminoacidato showed a better water solubility than the corresponding aromatic with exception of the aminobutiric acid derivative. All compounds showed an effective magnetic moment (1.68–2.05 BM) that agrees with paramagnetic species with one unpaired electron. The conductimetric measurements of ternary compounds shown a 1:1 type electrolyte (in water for compounds 1–8 and in methanol for compounds 9–12) confirming the +2 oxidation state of copper ion.
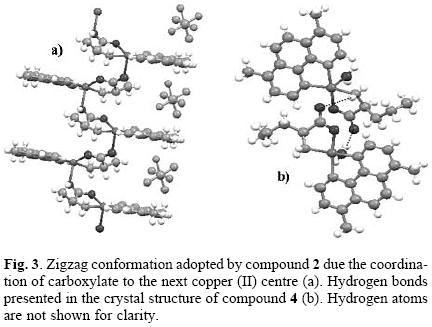
X–ray structure determination for ternary copper (II) com–plexes previously reported by our group [41–50], [Cu(H2O)(1,10–phenanthroline)(L–serinato)]NO3, [Cu(H2O)(3,4,7,8–tetramethyl–1,10–phenanthroline)(L–glycinato)]NO3 or [Cu(H2O)(4,7–diphenyl–1,10–phenanthroline)(L–alaninato)]NO3, present a slightly distorted square pyramid coordination environment around the copper ion with a water molecule occupying the apical position. Figure 2 shows the molecular structure of compounds 2 and 4. The molecular geometry of the cationic compound 2 (Cas IIala) is also a five coordinated Cu (II) centre but in an extremely distorted square–pyramidal geometry because the apical position is occupied by an oxygen atom of the carboxylic group of the neighboring coordination compound. Compound 4 (Cas IInval) presents the slightly distorted square base pyramid with the water molecule occupying the apical position already found in these copper (II) coordination compounds. Table 1 summarizes the crystallographic data for compounds 2 and 4. Selected bond lengths and angles are shown in Table 2.



The coordination of the oxygen in the carboxylate moiety to the copper ion in compound 2 promotes a zigzag conformation of the molecules in a 1D chain arrangement as shown in Figure 3. Hydrogen bond interactions also help in the stabilization of the crystal arrangement; a PF6– molecule interacts with hydrogen atoms of four different coordination compound molecules belonging of two 1D chains, with hydrogen bond distances as follows: F···H–NH–, 2.43(6) Å, F···H–CH2–, 2.56(3) Å, F···H–C(aromatic), 2.54(7) Å and F···H–C(aromatic), 2.65(4) Å. Also can be observed an intra–chain hydrogen bond formed between carboxylate oxygen of one molecule and the methylene hydrogen of the next compound. In compound 4, the coordinated water molecule and the non–coordinated oxygen of the carboxylate are involved in a hydrogen bond, the former interacts with the non–coordinated carboxylate oxygen of the nearby coordination compound (O···H–OH, 1.99(8) Å), meanwhile, the latter interacts with the hydrogens of the amino group of the same neighbor (O···H–NH–, 2.46(7) Å) (Fig. 3).
The antiproliferative effect of the compounds was determined as IC50 value on human tumour cell lines HeLa, HCT–15 and SKL–U by means of colorimetric microculture assay (sulforhodamine B assay) and reported in Table 3 as micromolar doses. Almost all compounds show lower doses than 10 μM on the three tumour cell lines with exception of compounds 2 with 13.96 μM in HCT–15 and 6 with 10.10 μM in SKL–U cell line.

In almost all cases, lower values than those exhibited by cisplatin in the different cell lines were found. Figure 4 shows the greater susceptibility of HeLa cells to the tested compounds, the IC50 values in this tumour cell line range from 1.3 μM, for compound 1, to 3.0 μM, for compound 11. This narrow interval reflects a lack of sensitivity to structural changes in aminoacidato side–chain on HeLa cell line. The same trend has been observed before in previous works where different cell lines exhibit slight susceptibility differences to structural changes [40]. The group with heterocyclic aminoacidates (group 2) is slightly more active than its counterparts with aliphatic aminoacidates (group 1), this trend is more clear on HCT–15 and SKL–U where the differences between values, as mentioned before, are greater than on HeLa.

Regarding group 1 (compounds 1–8), the structural difference lies in the length of the carbon side–chain, which brings simultaneously differences on steric effects and hydrophobicity. The latter property is a determining factor for drug transport across membranes. Cellular copper intake is tightly regulated by specific membrane transporter Ctr1 [51–53]; however, the change in hydrophobicity might facilitate the non regulated transport across membranes causing copper overload and increasing, as consequence, the biological response.
In order to understand the influence of hydrophobicity on copper uptake, the intracellular copper quantification by atomic absortion spectroscopy on HeLa cultures treated with 200 μM of complexes 1–8 by 15 min was performed. It is worthy to emphasize that the culture response to this concentration during 15 min exposure was evaluated before by Sulforhodamine B assay without changes on proliferation (data not shown). Results confirm that intracellular copper concentrations are increased in the cultures exposed to the complexes when compared with cultures exposed to Cu(NO3)2 (Fig. 5), leading to suggest that the presence of the ligands assist the copper uptake. The general trend indicates that the more hydrophobic the structure is, the more intracellular copper is found; however, the intracellular concentration does not strictly correlate with Log P and IC50 values.

The variations in the IC50 values are clearly related with structural changes, modifications on diimine ligand (primary ligand) promote greater changes in the in vitro antiproliferative activities than the modifications done in the aminoacidate side chain.
With the aim of confirming that modifications in the ligand produce minimal modifications in the antiproliferative activities of the Cu(II) coordination compounds, IC50 for groups 1 and 2 were plotted together with IC50 previously reported for complexes [Cu(N–N)(glycinate)]NO3 on HeLa cell line [40], where the structural changes were performed only on diimina ligand (Fig. 6). Since previous QSAR studies revealed both, the strong relationship of the biologic activity with the electronic environment of the metal center and the influence of N–N ligand basicity on copper electronic properties, pKa for diimina ligand (pKaN–N was selected as x axis to complete the comparisons. Figure 6 shows, as expected, that variations on diimine moiety have a stronger influence on antiproliferative activity, and are described adequately by pKa (r = –0.8548).

In this work we attempted to understand the influence of the secondary ligand in Cu (II) coordination compounds with general formula [Cu(4,7–dimethyl–1,10–phen)(N–O)]NO3 on the antiproliferative activity against human tumor cell lines HeLa, HCT–15 and SKL–U, employing a set of compounds with structural variations only on the side chain of aminoacidato. We found that the hydrophobicity of the ligands increases the cellular uptake of copper (II); however, the requirements of this property for in vitro activity are satisfied by all molecules and it is not the main limiting factor in the mode of action as it is demonstrated by the narrow interval of IC50 for this set of complexes, and confirmed by the lack of correlation of copper intracellular concentrations with in vitro activity. As reported before the substituents on diimine control the electronic environment around copper the atom [40] and this parameter is the dominant factor in the mode of action. The secondary ligand could have a key role in vivo where stability of ternary complexes and hydrophobicity are required to address the molecule to the final target.
Experimental
All chemicals and solvents were purchased from Aldrich Chemical Co. and GFS Chemicals Inc., and were used without further purification. Elemental analysis was done in a Fission Instruments Analyzer EA 1108, IR spectra were obtained employing a Nicolet Avatar 320 FT–IR, conductimetric and magnetic determinations were done in a Jenway 4330 Conductivity and pH meter and a Mkl magnetic balance from Sherwood Scientific respectively, UV–vis spectra were recorded in a Hewlett Packard 8452 diode array spectrophotometer. The UV–vis spectra and conductimetry were recorded in MeOH for some compounds (9–13) due to their low solubility in water.
Synthesis of [Cu(4,7–dimethyl–1,10–phenanthroline)(α–aminoacidato)(H2O)]NO3. All compounds were synthesized following the reported patents [25, 26].
Aqua (4,7–dimethyl–1,10–phenanthroline)(glycinato) copper(II) nitrate·hydrate (Casiopeína IIgly) (1). Yield 90%. Elemental analysis data: calculated (%) for C16H20CuN4O7 (443.9): C, 43.57; N, 12.52; H, 4.58. Found (%): C, 43.29; N, 12.62; H, 4.54. IR (KBr, ν/cm–1): 3305, 3253, 2945, 1600, 1579, 1430, 1383 (NO3–), 871, 726, 637. UV (H2O, ε/M–1cm–1) nm: λmax 611 (116). μeff = 1.78 BM. Λ (H2O) = 102.05 mS.
(4,7–dimethyl–1,10–phenanthroline)(alaninato) copper(II) nitrate (Casiopeína IIala) (2). Yield 70%. Elemental analysis data: calculated (%) for C17H18CuN4O5 (421.9): C, 48.24; N, 13.2; H, 4.28. Found (%): C, 48.4; N, 13.28; H, 4.3. IR (KBr, ν/cm–1): 3430, 2940, 1656, 1578, 1427, 1382 (NO3–), 1157, 932, 871, 726, 637. UV (H2O, ε/M–1cm–1) nm: λmax 596 (120). μeff = 1.895 BM. Λ (H2O) = 104.65 mS.
Aqua (4,7–dimethyl–1,10–phenanthroline)(valinato) copper(II) nitrate (Casiopeína IIval) (3). Yield 60%. Elemental analysis data: calculated (%) for C19H24CuN4O6 (467.95): C, 48.67; N, 11.98; H, 5.3. Found (%): C, 48.77; N, 11.97; H, 5.17. IR (KBr, ν/cm–1): 3305, 3243, 2961, 1647, 1578, 1427, 1385 (NO3–), 1130, 933, 871, 726, 637. UV (H2O, ε/M–1cm–1) nm: λmax = 609 (100). μeff = 1.814 BM. Λ (H2O) = 100.49 mS.
Aqua (4,7–dimethyl–1,10–phenanthroline)(norvalinat o) copper(II) nitrate (Casiopeína IInval) (4). Yield 10%. Elemental analysis data: calculated (%) for C19H26CuN4O7 (485.97): C, 46.95; N, 11.52; H, 5.39. Found (%): C, 47.35; N, 11.81; H, 5.38. IR (KBr, ν/cm–1): 3437, 3244, 2960, 1624, 1580, 1426, 1384 (NO3–), 1130, 932, 871, 726, 690. UV (H2O, ε/M–1cm–1) nm: λmax 602 (57). μeff = 1.754 BM. Λ (MeOH) = 91.3 mS.
Aqua (4,7–dimethyl–1,10–phenanthroline)(leucinato) copper(II) nitrate (Casiopeína IIleu) (5). Yield 50%. Elemental analysis data: calculated (%) for C20H26CuN4O6 (481.98): C, 49.63; N, 11.52; H, 5.44. Found (%): C, 49.84; N, 11.62; H, 5.44. IR (KBr, ν/cm–1): 3440, 3261, 2954, 1649, 1580, 1425, 1383 (NO3–), 1118, 931, 871, 726, 637. UV (H2O, ε/M–1cm–1) nm: λmax 595 (86). μeff = 1.771 BM. Λ (H2O) = 100.10 mS.
Aqua (4,7–dimethyl–1,10–phenanthroline)(isoleucinato) copper(II) nitrate·dihydrate (Casiopeína IIile) (6). Yield 60%. Elemental analysis data: calculated (%) for C20H30CuN4O8 (517.98): C, 46.42; N, 10.58; H, 6.02. Found (%): C, 46.37; N, 10.82; H, 5.84. IR (KBr, ν/cm–1): 3463, 2963, 1615, 1579, 1427, 1384 (NO3–), 1172, 930, 871, 726, 637. UV (H2O, ε/M–1cm–1) nm: λmax 605 (76). μeff = 1.685 BM. Λ (H2O) = 98.25 mS.
Aqua (4,7–dimethyl–1,10–phenanthroline)(norleucinato) copper(II) nitrate·hydrate (Casiopeína IInleu). (7) Yield 65%. Elemental analysis data: calculated (%) for C20H28CuN4O7 (499.98): C, 48.02; N, 11.44; H, 5.26. Found (%): C, 48.04; N, 11.21; H, 5.64. IR (KBr, ν/cm–1): 3313, 3243, 2954, 1657, 1579, 1426, 1384 (NO3–), 1116, 932, 871, 726, 637. UV (H2O, ε/M–1cm–1) nm: λmax 614 (50). μeff = 1.692 BM. Λ (H2O) = 126.88 mS.
Aqua (4,7–dimethyl–1,10–phenanthroline)(α–aminobutiric acid) copper(II) nitrate·dihydrate (Casiopeína IIaaba) (8). Yield 72%. Elemental analysis data: calculated (%) for C18H26CuN4O8 (489.93): C, 44.67; N, 12.19; H, 5.27. Found (%): C, 44.12; N, 11.43; H, 5.53. IR (KBr, ν/cm–1): 3326, 3222, 2962, 1627, 1580, 1423, 1383 (NO3–), 1160, 932, 871, 726, 637. UV (H2O, ε/M–1cm–1) nm: λmax 626 (50). μeff = 2.05 BM. Λ (H2O) = 121.55 mS.
Aqua (4,7–dimethyl–1,10–phenanthroline)(phenylalanin ato) copper(II) nitrate·hydrate (Casiopeína IIphe) (9). Yield 69%. Elemental analysis data: calculated (%) for C23H27CuN4O7 (535.04): C, 51.37; N, 10.47; H, 5.04. Found (%): C, 51.67; N, 10.49; H, 5.09. IR (KBr, ν/cm–1): 3423, 2945, 1622, 1579, 1525, 1427, 1383 (NO3–), 1137, 858, 726, 637. UV (MeOH, ε/M–1cm–1) nm: λmax 604 (70). μeff = 1.72 BM. Λ (MeOH) = 89.3 mS.
Aqua (4,7–dimethyl–1,10–phenanthroline)(prolinato) copper(II) nitrate·hydrate (Casiopeína IIpro) (10). Yield 63%. Elemental analysis data: calculated (%) for C19H22CuN4O6 (465.95): C, 48.83; N, 12.06; H, 4.86. Found (%): C, 49.02; N, 12.04; H, 4.77. IR (KBr, ν/cm–1): 3428, 2945, 1641, 1578, 1523, 1426, 1383 (NO3–), 1176, 852, 726, 637. UV (MeOH, ε/M–1cm–1) nm: λmax 598 (60). μeff = 1.70 BM. Λ (MeOH) = 90.2 mS.
(4,7–dimethyl–1,10–phenanthroline)(tyrosinato) copper(II) nitrate (Casiopeína IItyr) (11). Yield 60%. Elemental analysis data: calculated (%) for C23H22CuN4O6 (513.08): C, 53.74; N, 10.76; H, 4.47. Found (%): C, 53.76; N, 10.9; H, 4.31. IR (KBr, ν/cm–1): 3303, 2945, 1660, 1580, 1525, 1426, 1384 (NO3–), 1173, 864, 726, 637. UV (MeOH, ε/M–1cm–1) nm: λmax 602 (80). μeff = 1.71 BM. Λ (MeOH) = 80.2 mS.
Aqua (4,7–dimethyl–1,10–phenanthroline)(histidinato) copper(II) nitrate·hydrate (Casiopeína IIhis) (12). Yield 41%. Elemental analysis data: calculated (%) for C20H24CuN6O9 (523.10): C, 45.34; N, 16.24; H, 4.54. Found (%): C, 45.84; N, 16.04; H, 4.64. IR (KBr, ν/cm–1): 3385, 2945, 1629, 1578, 1525, 1425, 1383 (NO3–), 1172, 856, 726, 637. UV (MeOH, ε/M–1cm–1) nm: λmax 662 (95). μeff = 1.72 BM. Λ (MeOH) = 92.6 mS.
(4,7–dimethyl–1,10–phenanthroline)(tryptophanato) copper(II) nitrate (Casiopeína IItrp) (13). Yield 55%. Elemental analysis data: calculated (%) for C25H23CuN5O5 (537.03): C, 55.91; N, 13.16; H, 5.05. Found (%): C, 55.96; N, 13.06; H, 4.32. IR (KBr, ν/cm–1): 3423, 2945, 1626, 1580, 1526, 1427, 1384 (NO3–), 1173, 852, 726, 637. UV (MeOH, ε/M–1cm–1) nm: λmax 606 (70). μeff = 1.72 BM. Λ (MeOH) = 91.8 mS.
Tumor cell lines. HeLa (Cervix adenocarcinoma), HCT–15 (Colorectal adenocarcinoma) and SKL–U (Lung adenocarcinoma) cell lines were purchased from the American Type Culture Collection (ATCC) and propagated in Dulbecco's Modified Eagle Medium (D–MEM, Gibco Invitrogen Corporation) supplemented with 10% fetal bovine serum (FBS, Gibco Invitrogen Corporation).
Cell growth inhibition. Details of measuring cell growth inhibition are described elsewhere [54, 55]. Briefly, 2 × 104 cells/well were plated in 96–well microplate with Dulbecco's Modified Eagle Medium (D–MEM) supplemented with 10% fetal bovine serum (FBS), and allowed to attach by incubating at 37 °C and 5% CO2 for 24 h. At the end of incubation timing, the medium was aspired and cells were exposed to drugs in six different concentrations (0.1, 0.3, 1, 3, 10 and 30 μg/ml) for 24 h in the conditions mentioned above. Cisplatin (cisPt) was employed as control drug. Cell growth was determined according to the Sulforhodamine B assay [54, 55]. Absorbance was measured at 564 nm in a BIO–RAD 550 Microplate reader and % cell growth by each concentration of drug was calculated as: % growth = 100 * [T/C]; where T is the absorbance of treated wells and C is the absorbance of untreated wells. The 50% growth inhibition (IC50) was calculated by Probit survival analysis in StatPlus® 2005 [56]. Reported IC50 values are the average of at least three independent experiments.
Intracellular Copper quantification, 2 × 106 cells/well were cultured in a 12–well microplate with Dulbecco's Modified Eagle Medium (D–MEM) supplemented with 10% fetal bovine serum (FBS), and allowed to attach by incubating at 37 °C and 5% CO2 for 24 h. At the end of incubation time, the medium was aspired and cells were exposed to 200 μM of compounds 1–8 for 10 min in the conditions mentioned above. After exposition time, cells were washed three times with cold PBS pH 7.4 and recovered with lysis buffer pH 7.4 (NaCl 150mM, EDTA 1mM, SDS 0.1%, Triton X–100 1%, Tris–HCl 50mM, PMSF 1mM). Two aliquots were separated of each sample in order to determine copper concentration and total protein concentration; the former was determined with Graphite Furnace Atomic Absorption Spectrophotometry (Varian Spectr AA 220–Graphite Furnace GTA 110). Total protein was determined with DC BioRad's protein kit with a HP 8452A diode array spectrophotometer.
Single–crystal X–ray diffraction. Diffraction data for the complex was collected at 293 K on a Brucker 6000 CCD, using a monochromated Mo Kα radiation (λ = 0.71073 Å). The structure was solved by direct methods using SHELXS–97–2, least–squares refinement based on F2 was carried out by full–matrix method [57]. All non–hydrogen atoms were refined with anisotropic thermal parameters. The hydrogen atoms for all the reported structures were located in the difference map and included in the refinement with an isotropic fixed thermal parameter using a "riding" model. Neutral atom scattering factors and anomalous dispersion corrections were obtained from the International Tables for Crystallography vol. A [58]. Unit cell parameters along with data collection and refinement details for these complexes are listed in Table 1. All molecular structure drawings were generated using the WINGX suite of crystallographic programs for Windows [59].
Acknowledgments
To CONACyT scholarship (SDM), SNI (JCGR), PAPIIT (MAMH) and F–634–3 (JFG). CONACyT–Salud 87806 and PAPIIT IN227110.
Supporting Information Available
The crystallographic data can be obtained free of charge via www.ccdc.cam.ac.uk/data_request/cif, or by emailing data_request@ccdc.cam.ac.uk, or by contacting The Cambridge Crys–tallographic Data Centre, 12, Union Road, Cambridge CB2 1EZ, UK; fax +44 1223 336033. CCDC 826177 , 826178 contains the supplementary crystallographic data for compounds 4 and 2, respectively.
References
1. Hambley, T. W. Science 2007, 318, 1392–1393. [ Links ]
2. Zhang, C. X.; Lippard, S. J. Curr. Opin. Chem. Biol. 2003, 7, 481–489. [ Links ]
3. van Rijt, S. H.; Sadler, P. J. Drug Discov. Today 2009, 14, 1089–1097. [ Links ]
4. Mejía–Vázquez, M. d. C.; Navarro, S. New Approaches in the Treatment of Cancer, Cancer Etiology, Diagnosis and Treatments Nova Sciences, New York, Chapter 10, 2010. [ Links ]
5. Alama, A; Tasso, B; Novelli, F. ; Sparatore, F. Drug Discov. Today 2009, 14, 500–508. [ Links ]
6. Kostova, I. Anticancer Agents Med. Chem. 2009, 9, 827–842. [ Links ]
7. Messori, L.; Marcon, G.; Orioli, P. Bioinorg. Chem. Appl. 2003, 177–187. [ Links ]
8. Amin, A.; Buratovich, M. A. Mini Rev. Med. Chem. 2009, 9, 1489–1503. [ Links ]
9. Gao, E.; Liu, C.; Zhu, M.; Lin, H.; Wu, Q.; Liu, L. Anticancer Agents Med. Chem. 2009, 9, 356–368. [ Links ]
10. Chifotides, H. T.; Dunbar, K. R. Acc. Chem. Res. 2005, 38, 146–156. [ Links ]
11. Hindo, S. S.; Frezza, M.; Tomco, D.; Heeg, M. J.; Hryhorczuk, L.; McGarvey, B. R.; Dou, Q. P.; Verani, C. N. Eur. J. Med. Chem. 2009, 44, 4353–4361. [ Links ]
12. Marzano, C.; Pellei, M; Tisato, F.; Santini, C. Anticancer Agents Med. Chem. 2009, 9, 185–211. [ Links ]
13. Ruiz–Azuara, L.; Bravo–Gomez, M. E. Curr. Med. Chem. 2010, 17, 3606–3615. [ Links ]
14. Tardito, S.; Marchio, L. Curr. Med. Chem. 2009, 16 (2009) 1325–1348. [ Links ]
15. Tisato, F.; Marzano, C.; Porchia, M.; Pellei, M.; Santini, C. Med. Res. Rev. 2010, 30, 708–749. [ Links ]
16. Fraústo da Silva, J. J. R.; Williams, R. J. P. The Biological Chemistry of the Elements – The Inorganic Chemistry of Life, Oxford University Press, Oxford, 1991. [ Links ]
17. Olivares, M.; Uauy, R. Am. J. Clin. Nutr. 1996, 63, 791S–796S. [ Links ]
18. Halliwell, B.; Gutteridge, J. M. Methods Enzymol. 1990, 186, 1–85. [ Links ]
19. Gaetke, L. M.; Chow, C. K. Toxicology 2003, 189, 147–163. [ Links ]
20. Hill, R. Br. Vet. J. 1977, 133, 365–373. [ Links ]
21. Hill, R. Br. Vet. J. 1977, 133, 219–224. [ Links ]
22. Miller, D. M.; Buettner, G. R.; Aust, S. D. Free Radic. Biol. Med. 1990, 8, 95–108. [ Links ]
23. Sandstead, H. H. Am. J. Clin. Nutr. 1995, 61, 621S–624S. [ Links ]
24. Ruiz–Azuara, L. US Patent 1992 Re 35, 458. [ Links ]
25. Ruiz–Azuara, L. US Patent 1992 RE 35, 458, 1997. [ Links ]
26. Ruiz–Azuara, L. US Patent 1996 5, 576, 326. [ Links ]
27. Alemón–Medina, R.; Breña–Valle, M.; Muñoz–Sánchez, J. L.; Gracia–Mora, M. I.; Ruiz–Azuara, L. Cancer Chemother. Pharmacol. 2007, 60, 219–228. [ Links ]
28. Gracia–Mora, I.; Ruiz–Ramírez, L.; Gómez–Ruiz, C.; Tinoco–Méndez, M.; Márquez–Quiñones, A.; Romero–De Lira, L.; Marín–Hernández, A.; Macías–Rosales, L.; Bravo–Gómez, M.E. Metal–Based Drugs 2001, 8, 19–28. [ Links ]
29. Mejia, C.; Ruiz–Azuara, L. Pathol. Oncol. Res. 2008, 14, 467–472. [ Links ]
30. Carvallo–Chaigneau, F.; Trejo–Solís, C.; Gómez–Ruiz, C.; Rodríguez–Aguilera, E.; Macías–Rosales, L.; Cortés–Barberena, E.; Cedillo–Peláez, C.; Gracia–Mora, I.; Ruiz–Azuara, L. ; Madrid–Marina, V.; Constantino–Casas, F. Biometals 2008, 21, 17–28. [ Links ]
31. Trejo–Solís, C.; Palencia, G.; Zuniga, S.; Rodriguez–Ropon, A.; Osorio–Rico, L.; Luvia, S. T.; Gracia–Mora, I.; Márquez–Rosado, L.; Sánchez, A.; Moreno–García, M. E.; Cruz, A.; Bravo–Gómez, M. E.; Ruiz–Ramírez, L.; Rodríguez–Enríquez, S.; Sotelo, J. Neoplasia 2005, 7, 563–574. [ Links ]
32. De Vizcaya–Ruiz, A.; Rivero–Muller, A.; Ruiz–Ramirez, L.; Kass, G. E.; Kelland, L. R.; Orr, R. M.; Dobrota, M. Toxicol. In Vitro 2000, 14, 1–5. [ Links ]
33. Alemón–Medina, R.; Muñoz–Sánchez, J. L.; Ruiz–Azuara, L.; Gracia–Mora, I. Toxicol. In Vitro 2008, 22, 710–715. [ Links ]
34. Rivero–Muller, A.; De Vizcaya–Ruiz, A.; Plant, N.; Ruiz, L.; Dobrota, M. Chem. Biol. Interact. 2007, 165, 189–199. [ Links ]
35. Hernández–Esquivel, L.; Marín–Hernández, A.; Pavón, N.; Carvajal, K.; Moreno–Sánchez, R. Toxicol. Appl. Pharmacol. 2006, 212, 79–88. [ Links ]
36. Marín–Hernández, A. Gracia–Mora, I. Ruiz–Ramírez, L. Moreno–Sánchez, R. Biochem. Pharmacol. 2003, 65, 1979–1989. [ Links ]
37. Ruili Huang, A. W.; Covell, D. G. Biochem. Pharmacol. 2005, 69, 1009–1039. [ Links ]
38. Sigman, D. S.; Mazumder, A.; Perrin, D. M. Chem. Rev. 1993, 93, 2295–2316. [ Links ]
39. Chikira, M.; Tomizawa, Y.; Fukita, D.; Sugizaki, T.; Sugawara, N.; Yamazaki, T.; Sasano, A.; Shindo, H.; Palaniandavar, M.; Antholine, W. E. J. Inorg. Biochem. 2002, 89, 163–173. [ Links ]
40. Bravo–Gómez, M. E.; García–Ramos, J. C.; Gracia–Mora, I.; Ruiz–Azuara, L. J. Inorg. Biochem. 2009, 103, 299–309. [ Links ]
41. Tovar–Tovar, A.; Ruiz–Ramírez, L.; Campero, A.; Romerosa, A.; Moreno–Esparza, R.; Rosales–Hoz, M. J. J. Inorg. Biochem. 2004, 98, 1045–1053. [ Links ]
42. Solans, X.; Ruiz–Ramírez, L.; Gasque, L.; Briansó, J. L. Acta Cryst. C 1987, 43, 428–430. [ Links ]
43. Solans, X.; Ruiz–Ramírez, L.; Martínez, A.; Gasque, L.; Briansó, J. L. Acta Cryst. C 1988, 44, 628–631. [ Links ]
44. Solans, X.; Ruiz–Ramírez, L.; Martínez, A.; Gasque, Moreno–Esparza, R. Acta Cryst. C 1992, 48, 1785–1788 [ Links ]
45. Solans, X.; Ruiz–Ramírez, L.; Martínez, A.; Gasque, Moreno–Esparza, R. Acta Cryst C 1993, 49, 890–893. [ Links ]
46. Moreno–Esparza, R.; Molins, E.; Briansó–Penalva, J. L.; Ruiz–Ramírez, L.; Redón, R. Acta Cryst C 1995, 51, 1505–1508. [ Links ]
47. Gasque, L.; Moreno–Esparza, R.; Ruiz–Ramírez, L.; Medina–Dickinson, G. Acta Cryst C 1999, 55, 1065–1067. [ Links ]
48. Gasque, L.; Moreno–Esparza, R.; Ruiz–Ramírez, L.; Medina–Dickinson, G. Acta Cryst C 1999, 55, 1063–1065. [ Links ]
49. Alvarez–Larena, A.; Briansó–Penalva, J. L.; Piniella, J. F.; Moreno–Esparza, R.; Ruiz–Ramírez, L.; Ferrer–Sueta, G. Acta Cryst. C 1995, 51, 852–854. [ Links ]
50. Paulovicova, A; El–Ayaan, U.; Fukuda, Y. Inorg. Chim. Acta 2001, 321, 56–62. [ Links ]
51. Huffman, D. L.; O'Halloran, T. V.; Annu. Rev. Biochem. 2001, 70, 677–701. [ Links ]
52. Lee, J.; Pena, M. M.; Nose, Y.; Thiele, D. J. J. Biol. Chem. 2007, 277, 380–4387. [ Links ]
53. Markossian, K. A.; Kurganov, B. I. Biochemistry (Moscow) 2003, 68, 827–837. [ Links ]
54. Rubinstein, L. V.; Shoemaker, R. H.; Paull, K. D.; Simon, R. M.; Tosini, S.; Skehan, P.; Scudiero, D. A.; Monks, A.; Boyd, M. R. J. Natl. Cancer. Inst. 1990, 82, 1113–1118. [ Links ]
55. Skehan, P.; Storeng, R.; Scudiero, D.; Monks, A.; McMahon; Vistica, D.; Warren, J. T.; Bokesch, H.; Kenney, S.; Boyd, M. R. J. Natl. Cancer. Inst. 1990, 82, 1107–1112. [ Links ]
56. AnalystSoft, Robust Business Solutions, StatPlus 2005 Professional Build Version 3.5.3.0, 2005. [ Links ]
57. SHELX–97–2 [Includes SHELXS97, SHELXL97, CIFTAB (and SHELXA)] – Programs for Crystal Structure Analysis (Release 97–2). Sheldrick, G. M., Institüt für Anorganische Chemie der Universität, Tammanstrasse 4, D–3400 Göttingen, Germany, 1998. [ Links ]
58. INTERNATIONAL TABLES VOL. A – Hahn, T. Ed., International Tables for Crystallography, Volume A, Kluwer Academic Publishers, Dordrecht, The Netherlands, 1995. [ Links ]
59. L. J. Farrugia, J. Appl. Cryst., 1999, 32, 837. [ Links ]
Note
Dedicated to Dr. Estela Sánchez de Jiménez for her invaluable contributions to plant biochemistry.














