Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Journal of the Mexican Chemical Society
versión impresa ISSN 1870-249X
J. Mex. Chem. Soc vol.56 no.1 Ciudad de México ene./mar. 2012
Article
Purification and Characterization of an Alkaline Phosphatase Induced by Phosphorus Starvation in Common Bean (Phaseolus vulgaris L.) Roots
Lorena Morales, Natalia Gutiérrez, Vanessa Maya, Carmen Parra, Eleazar Martínez–Barajas, and Patricia Coello*
Departamento de Bioquímica, Facultad de Química. Universidad Nacional Autónoma de México. México, 04510 D.F. 55–56–22–53–29; pcoello@servidor.unam.mx
Received January 6, 2011.
Accepted April 25, 2011.
Abstract
Two phosphatase isoforms from roots of the common bean (Phaseolus vulgaris L.) showed an increase in activity in response to phosphate deficiency. One of them (APIII) was chosen for further purification through ionic exchange chromatography and preparative electrophoresis. The estimated molecular mass of APIII was 35 kDa by both SDS–PAGE and gel filtration analyses, suggesting a monomeric form of the active enzyme. The phosphatase was classified as an alkaline phosphatase based on the requirement of pH 8 for optimum catalysis. It not only exhibited broad substrate specificity, with the most activity against pyrophosphate, but also effectively catalyzed the hydrolysis of polyphosphate, glucose–1–phosphate and phosphoenol–pyruvate. Activity was completely inhibited by molybdate, vanadate and phosphate but was only partially inhibited by fluoride. Although divalent cations were not essential for the pyrophosphatase activity of this enzyme, the hydrolysis of pyrophosphate increased substantially in the presence of Mg2+.
Key words: Alkaline phosphatase, Phaseolus vulgaris, phosphate deficiency, pyrophosphate, purification.
Resumen
Dos isoformas de fosfatasas obtenidas de raíz de frijol (Phaseolus vulgaris L.) mostraron un incremento en la actividad en respuesta a la deficiencia de fosfato. Una de ellas (APIII) se purificó a través de una cromatografía de intercambio iónico y una electroforesis preparativa. La masa molecular estimada para APIII fue de 35 kDa tanto por SDS–PAGE como por filtración molecular, sugiriendo que la enzima activa es monomérica. APIII se clasificó como una fosfatasa alcalina basada en sus requerimientos de pH 8 para catálisis. Esta enzima es activa sobre un amplio espectro de sustratos como polifosfato, glucose 1–fosfato y fosfoenolpiruvato, aunque muestra preferencia por pirofosfato. Su actividad se inhibe completamente por molibdato, vanadato y fosfato, aunque es inhibida parcialmente por fluoruro. Aún cuando los cationes divalentes no fueron escenciales para su actividad, la hidrólisis de pirofosfato se incrementó notablemente en presencia de Mg2+.
Palabras clave: Fosfatasa alcalina, Phaseolus vulgaris, deficiencia de fosfato, pirofosfato, purificación.
Introduction
Phosphorus (P) is one of the major elements found in plants. The available phosphate concentration in many soils is rather low, in part because it is commonly bound to many soil constituents [1]. As the sources of P run out, improvements in the absorption and/or use–efficiency of phosphorus become necessary. Plants have developed several mechanisms to overcome P deficiency, such as induction of extracellular and intracellular phosphatases (ortophosphoric–monoester phosphohydrolase, EC 3.1.3.2) [2, 3 4, 5, 6,7]. Considering that the organic phosphorus content can reach up to 80% in some soils, extracellular acid phosphatases (ACP) are thought to play an important role in scavenging phosphate (Pi) from organic soil constituents. Similarly, intracellular phosphatases may be involved in regulating the supply of Pi from intracellular organic sources [8, 9]. Phosphatases have been traditionally classified as being alkaline or acid according to their optimal pH for catalysis [10]. Plant alkaline phosphatases generally display substrate specificity and play defined roles in metabolism. In contrast, acid phosphatases generally are nonspecific with the exception of PEP phosphatase [11]. Most studies on the relationship between Pi deprivation and phosphatases have focused on acid phosphatases.
The common bean (Phaseolus vulgaris) is a major food leguminous crop, mainly cultivated in P–deficient soils in the tropics [12]. Several groups have elucidated the morphological, physiological and genetic mechanisms underlying P–de–ficiency responses [13, 14, 15,16]. Phosphatase secretion has been related to the capacity to tolerate P deficiency in several plant species including P. vulgaris [17,18,19, 20]. In an effort to elucidate the importance of these phosphatases, we further purified and characterized one of the root isoforms induced by Pi starvation.
Results and Discussion
The common bean, like other plant species, responds to Pi starvation with an increase in the activity of acid phosphatases in roots and leaves [6, 20, 21]. Although it seems that leaf isoforms are not advantageous in low–phosphorus availability [22], the importance of root phosphatases has not been documented. In this study, we further characterized one of the root isoforms, an essential step in understanding its possible role in the Pi–deficiency response. Our results revealed two new phosphatase isoforms after a Pi–deficiency treatment (Fig. 1). The de novo synthesis of phosphohydrolases has been demonstrated in cells of Br8ssic8 nigr8 [9], in tomato seedlings [23] and white lupin roots [4], but the increase in phosphatase activity could also be due to the activation of pre–existing protein.
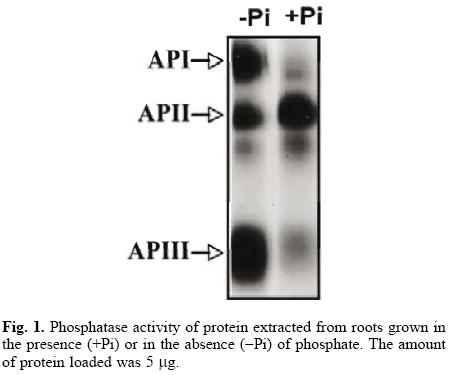
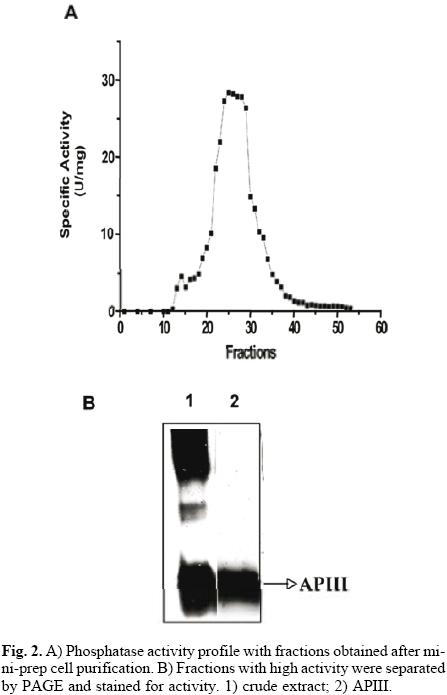
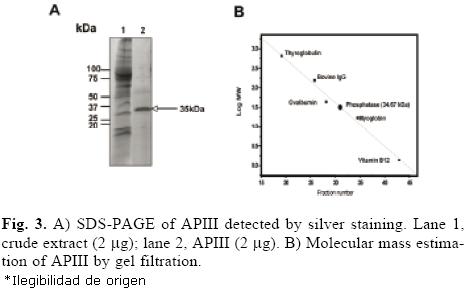
The protocol for the purification of root phosphatases consisted of two steps. The root extract was first loaded onto a DEAE–Sephacel column, and the proteins were eluted with a pH gradient from 4 to 8. The API isoform did not bind to the column (data not shown). The APII and APIII phosphatases eluted within the pH range of 4 to 5. Fractions with activity were pooled, concentrated and loaded onto a mini–preparative cell. A single peak of activity was eluted from the gel in fractions 24 to 30 (Fig. 2A). In–gel activity of these fractions showed that only APIII was present (Fig. 2B). SDS–PAGE analysis of APIII indicated that this enzyme is a protein composed of a single 35–kDa subunit (Fig. 3A). The molecular mass, estimated by gel filtration, of APIII was also 35 kDa, suggesting that the enzyme is active as a monomer (Fig. 3B). Thus far, most of the acid phosphatases that respond to Pi–deficiency are enzymes composed of 1 or 2 subunits with native molecular masses ranging between 50 kDa and 200 kDa [2, 3, 6, 24].
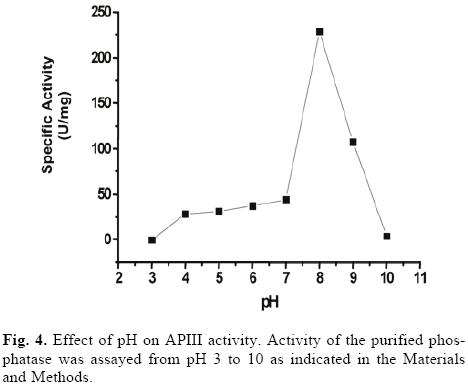
The optimum pH for catalysis by the purified phosphatase was 8, which classified it as an alkaline phosphatase (Fig. 4). To examine the pH stability of the enzyme, the purified phosphatase was incubated for 10 min at different pHs. No significant loss of activity was observed at pH 6–7, but 50% of the activity was lost at pH 5 and 9 (data not shown). With the exception of fructose 1,6 bi–phosphatase, sucrose 6– phosphate phosphatase [5] and an inducible alkaline phosphatase from Spirodela oligorrhiza [24], very few alkaline phosphatases are induced by Pi–deficiency. Plant alkaline phosphatases generally display rather strict substrate specificity and play defined roles in metabolism [10].
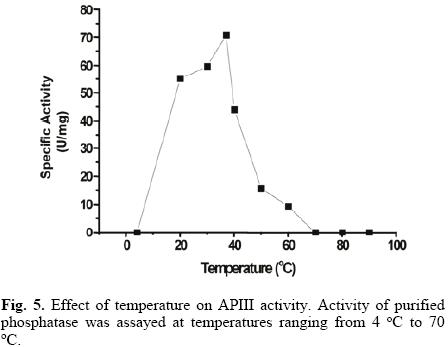
APIII activity strongly increased with temperature from 20 °C to 37 °C (Fig. 5). The presence of 20 mM of p–NPP reinforced enzyme thermal stability, with no loss of activity observed after 10 minutes, even at 70 °C (data not shown). Heat stable acid phosphatases have been reported in some plant species [25, 26], but to the best of our knowledge, the stability of plant alkaline phosphatases has not been documented.
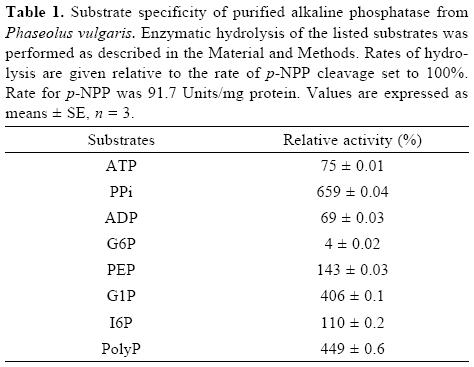
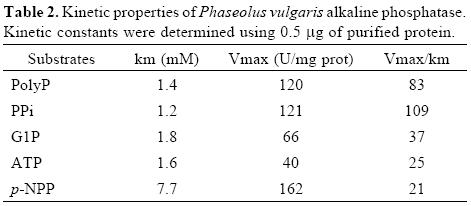
Although the alkaline phosphatase reported here showed relatively broad substrate specificity, the highest specificity constant was found with PPi as substrate (Table I and II). This enzyme, however, is not a typical pyrophosphatase because Mg2+ is not essential for its activity. Additionally, Km values for PPi are in the micromolar range for typical pyrophospha–tases [24, 27], whereas the value found in this work for APIII is in the order of millimolar. In plant cells, during Pi–deficiency, levels of cytoplasmic Pi as well as of nucleoside phosphates, such as ATP and ADP, decrease. In contrast, levels of pyrophosphate appear to remain more constant and may serve as a substitute energy donor [5]. Thus, this alkaline phosphatase, which exhibits high specificity toward PPi hydrolysis, might be an important intracellular enzyme to increase levels of Pi during Pi–deficiency.
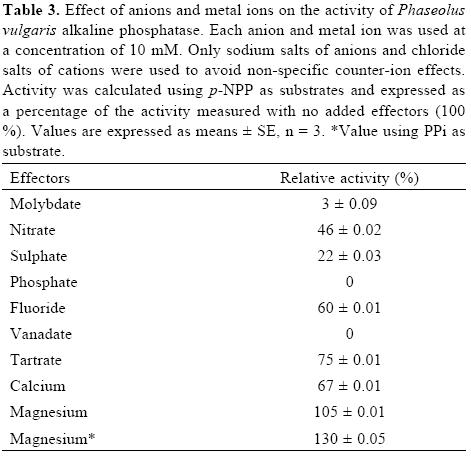
Molybdate, vanadate and phosphate are potent inhibitors of acid and alkaline phosphatases [2, 3]. In contrast, fluoride strongly inhibits the acid forms [3], but has a variable effect on alkaline phosphatases [25]. In agreement with the above, molybdate, vanadate and phosphate were strong inhibitors of APIII activity, but fluoride inhibited its activity only 40% (Table III). In the case of the S. oligorrhiza alkaline phosphatase, EDTA and EGTA do not inhibit its activity, suggesting that the enzyme does not depend on divalent metal ions [24]. The activity of the common bean APIII was enhanced 30% by Mg2+, whereas Ca2+ showed a slight inhibition.
Experimental section
Plant materials
Phaseolus vulgaris L. var. MAR1 seeds were disinfected for 1 min in a 10% (v/v) solution of NaOCl and thoroughly washed with distilled water. After 5 d of germination on wet paper, they were transferred to agrolite and watered daily with a complete nutritive Hoagland II solution (3 mM KNO3, 2 mM Ca (NO3)2, 1 mM MgSO4, 0.004 mM MnCI2, 0.023 mM H3BO3, 0.004 mM ZnSO4, 0.00015 mM CuSO4, 0.00005 mM H2MoO4 and 1 g/200 mL Fe (III) EDTA). The nutritive solutions differed only in the concentrations of Pi: for the Pi sufficiency (+Pi) treatments, a concentration of 500 uM ammonium phosphate was used, and for the Pi absence (–Pi) treatments, a solution with 500 μM ammonium sulfate was utilized. The plants were maintained in a greenhouse with cycles of 26 °C during the day and 15 °C at night with constant light. The relative humidity was maintained at 70%. The plants were watered with the Hoagland II complete nutritive solution for 10 d; at the two leaves stage, they were divided and treated with the respective nutritive solution, with or without Pi, for three more weeks.
Enzymatic extraction
To measure the enzymatic activities in root extracts, samples (5 g) were homogenized in a mortar at 4 °C with 1 vol of buffer containing 20 mM Tris–HCI pH 8.8, 150 mM NaCI, 1 mM EDTA, 20% (v/v) glycerol, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 μg/ml leupeptin and 1 mM benzamidine. The homogenized sample was centrifuged at 6,000 x g for 15 min, and the supernatant was used as an enzyme source.
Enzymatic assays
The acid phosphatase activity was measured using p–nitrophe–nyl phosphate (p–NPP, Sigma) as a substrate [3]. The enzyme was incubated with 20 mM p–NPP in a final volume of 0.5 ml, for 10 min at 37 °C, with a buffer containing 100 mM sodium acetate pH 4.8 or 100 mM Tris–HCl pH 8, 3 mM MgCI2, 1 mM EDTA and 10% (v/v) glycerol. The reaction was stopped by adding 1 ml of NaOH 1 M. The liberation of p–nitrophenol was measured by determining the absorption of the mixture at 410 nm in an Ultrospec 2000 (Pharmacia Biotech) spectropho–tometer. The amount of p–nitrophenol produced was calculated using a molar extinction coefficient of 20,000 M–1cm–1. A unit of phosphatase activity is defined as the amount of enzyme that liberates 1 mmol of Pi per minute. The optimum pH was measured with acetate buffer (0.1 M) in the pH range of 3–5, Mes–NaOH (0.1 M) in the range of 6–7 and Tris–HCl (0.1 M) in the range of 8–9.
With other substrates, enzyme activity was assayed by the amount of phosphate released [28]. After 10 min at 37°C with the corresponding substrate, the reaction was terminated by adding 0.7 ml of reagent A (1.4% ascorbic acid, 0.36% ammonium molybdate in 1 N H2SO4), followed by incubation for 20 min at 45 °C. The absorbance was measured at 829 nm. A standard curve was constructed for 0.05 mM to 1 mM KH2PO4 range.
For determination of catalytic parameters such as Km and Vmax for various substrates, the concentration ranging from 0.05 to 3 mM was used for PPi, G1P, and polyP, ADP. Additionally, p–NPP was used from 0.5 to 60 mM. When the influence of various substrates as activators or inhibitors was evaluated, the assay conditions did not include magnesium.
Analysis of the isoforms of acid phosphatase
The proteins were separated in a non–denaturing polyacryl–amide gel (PAGE) at 4 °C [29]. When the electrophoresis was completed, the gels were washed three times in 100 mM of sodium acetate, pH 5.0. The activity was visualized by means of the Fast Garnet method using 1 mM a–naphthyl phosphate as a substrate and 1 mM Fast Garnet as a dye in 100 mM of sodium acetate, pH 5 [3]. A dark colored precipitate indicated phosphatase activity. The reaction finished and the gels were fixed in a 1:5:5 ratio of acetic acid: methanol: water.
Protein purification
To isolate the phosphatase, crude extract from root plants, obtained as described before, was loaded onto a DEAE–Sephacel column (30 cm × 2 cm), previously equilibrated in buffer A (20 mM Tris–HCl pH 8, 1 mM EDTA, 3 mM MgCl2 and 10% glycerol (v/v)). Proteins were eluted using a pH gradient with buffer B (20 mM acetate pH 4, 1 mM EDTA, 3 mM MgCl2 and 10% glycerol (v/v)) and buffer A. Fractions with phosphatase activity were pooled, concentrated and loaded onto a mini–preparative cell (BIORAD). A 10% polyacrylamide resolving gel and a 4% polyacrylamide stacking gel were cast in the 7–mm gel tube of the mini prep cell. Non–denaturing PAGE was carried out in a discontinuous electrophoresis buffer system. The gel ran for 12 h at 1 W constant. Proteins were eluted with 10 mM Tris–HCl pH 8, and fractions of 1 mL were collected. Fractions with phosphatase activity were analyzed by SDS–PAGE and used for further characterization.
Molecular mass calculation
To estimate the molecular mass of the phosphatase, a Superdex–200 HR 10/30 column was used. The column was equilibrated with buffer containing 20 mM Tris–HCl pH 8, 3 mM MgCl2, 1 mM EDTA and 10% (v/v) glycerol. Proteins were eluted with the same buffer. Molecular markers used for the calibration were thyroglobulin (670 kDa), bovine IgG (158 kDa), ovalbumin (44 kDa), myoglobin (17 kDa) and vitamin B12 (1.35 kDa).
Determination of protein
Protein content was quantified by means of the Bradford method [30] using bovine serum albumin for the standard curve.
Acknowledgements
We thank Carlos Mújica and Laurel Fabila for technical support. This work was supported by PAPIIT–UNAM (grant IN205109), CONACyT 80038 and PAIP 6290–13.
References
1. Bieleski, R. L. Ann. Rev. Plant Physiol. 1973, 24, 225–252. [ Links ]
2. Bozzo, G. G.; Raghothama, K. G.; Plaxton, W. C. Eur. J. Biochem. 2002, 269, 6278–6286. [ Links ]
3. Coello, P. Physiol. Plant. 2002, 116, 293–298. [ Links ]
4. Gilbert, G. A. J. D. Knight, C. P. Vance, D. L. Allan. Plant Cell Env. 1999, 22, 801–810. [ Links ]
5. Poirier, Y.; Bucher, M. The Arabidopsis Book. American Society of Plant Biologist 2002, pp 1–35. [ Links ]
6. N. A. Tejera–García, N. A.; Olivera, N. M.; Iribarne, C.; Lluch, C. Plant Physiol and Biochem. 2004, 42, 585–591. [ Links ]
7. Liang, C.; Tian J.; Lam H. M.; Lim B. L.; Yan X.; Liao H. Plant Physiol. 2010, 152, 854–865. [ Links ]
8. M. Li, M.; Osaki, M.; IRao, I. M.; Tadano, T. Plant and soil 1997, 195, 161–169. [ Links ]
9. Trull, M. C.; Deikman, J. Planta 1998, 206, 544–550. [ Links ]
10. Duff , S. M. G.; Sarath, G.; Plaxton, W. C. Physiol Plant 1994, 90, 791–800. [ Links ]
11. Duff, S. M. G..; Moorhead, G. B. G.; Lefebvre, D. D.; Plaxton, W.C. Plant Physiol. 1989, 90, 1275–1278. [ Links ]
12. Beebe, S. E.; Rojas–Pierce, M.; Yan, X.; Blair, M. W.; Pedraza, F.; Muñoz, F.; Tohme, J.; Lynch, J. P. Crop Sci. 2006, 46, 413–423. [ Links ]
13. Shen, H.; Yan, X.; Zhao, M.; Zheng, S.; Wang, X. Environ Exp. Bot. 2002 48, 1–9. [ Links ]
14. Bernal, L.; Coello, P.; Acosta, J.; Martínez–Barajas, E. Agrociencia 2007, 41, 417–423. [ Links ]
15. Ramírez, M.; Graham, M. A.; Blanco–López, L.; Silvente, S.; Medrano–Soto, A.; Blair, M. W.; Hernández, G.; Vance, C. P.; Lara, M. Plant Physiol. 2005, 137, 1211–1227. [ Links ]
16. Graham, M. A.; Ramírez, M.; Váldes–López, O.; Larab, M.; Tesfaye, M.; Vance, C. P.; Hernández, G. Funct Plant Biol 2006, 33, 789–797. [ Links ]
17. Asmar, F.; Gahoonia, T.; Nielsen, N. Plant Soil 1995, 172, 117–122. [ Links ]
18. Gaume, A. F., Machler, F.; De León, C.; Narro, L.; Frossard, E. Plant and soil 2001, 228, 253–264. [ Links ]
19. Helal, H. M. Plant Soil 1990, 123,161–163. [ Links ]
20. Parra, C..; Martínez–Barajas, E.; Acosta, J.; Coello, P. Agrociencia 2004, 38, 131–139. [ Links ]
21. Yun, S. J.; Kaeppler, S. M. Plant and Soil 2001, 237, 109–115. [ Links ]
22. Yan, X.; Liao, H.; Trull, M. C.; Beebe, S. E.; Lynch, J. P. Plant Physiol. 2001, 125, 1901–1911. [ Links ]
23. Bosse, D.; Kock, M. Plant Cell Environ 1998, 21, 325–332. [ Links ]
24. Nakazato, H.; Okamoto, T.; Ishikawa, K.; Okuyama, H. Plant Physiol. Biochem. 1997, 35, 437–446. [ Links ]
25. Li, M.; Tadano, T. Soil Sci. Plant Nutr. 1996, 42, 753–763. [ Links ]
26. Turner, W.; Plaxton, W. C. Planta 2001, 214, 243–249. [ Links ]
27. Olczak, M.; KobiaIka, M.; Watorek, W, Biochim. Biophys. Acta 2000, 1478, 239–247. [ Links ]
28. Ames, B. N. Meth. Enzymol. 1966, 8,115–118. [ Links ]
29. Laemmli, U. K. Nature 1970, 227, 680–685. [ Links ]
30. Bradford, M. M. Anal Biochem. 1976, 72, 248–254. [ Links ]
Note
Dedicated to Dr. Estela Sánchez de Jiménez for her invaluable contributions to plant biochemistrys.














