Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Journal of the Mexican Chemical Society
versión impresa ISSN 1870-249X
J. Mex. Chem. Soc vol.56 no.1 Ciudad de México ene./mar. 2012
Article
Identification of Proteins from Cap–Binding Complexes by Mass Spectrometry During Maize (Zea mays L.) Germination
Pedro E. Lázaro–Mixteco and Tzvetanka D. Dinkova*
Departamento de Bioquímica, Facultad de Química, Universidad Nacional Autónoma de México, Ciudad Universitaria, Coyoacán 04510, México D.F. +52 55 56225277, cesy@unam.mx
Received January 5, 2011.
Accepted May 30, 2011.
Abstract
This work describes the identification of components in the Cap–binding complexes in non–germinated and 24–h–imbibed seeds using mass spectrometry. This approach revealed new components particularly present in the non–germinated seed. Among these, two heat shock proteins, HSP101 and HSP70, were detected as well as several proteins involved in carbohydrate metabolism. Between the new components of maice Cap–binding complexes, several proteins contain a motif that identifies them as potential direct interactors with eIF4E or eIF(iso)4E.Together with the major abundance of eIF(iso)4E at this developmental stage, our findings indicate clear differences between the translation initiation complexes that are available for protein synthesis right upon water imibition and those that are present once germination has been completed.
Key words: Cap–binding proteins, germination, eIF4E–binding motif, translation, zea mays L.
Resumen
En el presente trabajo se describe la identificación de componentes de los complejos de unión a Cap obtenidos de semillas de maíc sin germinar y embebidas en agua por 24 h utilicando espectrometría de masas. Mediante este procedimiento se encontraron nuevos componentes en los complejos de unión a Cap, presentes particularmente en las semillas sin germinar. Entre estos, se detectaron dos proteínas de choque térmico o chaperonas, HSP101 y HSP70, así como varias proteínas involucradas en metabolismo de carbohidratos. Entre los nuevos componentes de complejos de unión a Cap en maíc, varias proteínas presentan motivos de aminoácidos que los identifican como interactores directos potenciales de las proteínas eIF4E y eIF (iso)4E. Estos hallacgos indican que los complejos de inicio de la traducción difieren entre el inicio y término de la germinación de semillas de maíc, tanto por la abundancia de eIF(iso)4E como por su composición proteica.
Palabras clave: Germinación, motivo de unión a EiF4E, proteínas de unión a cap, traducción, zea mays L.
Introduction
Translation of mRNAs in eukaryotes initiates through their 5' end Cap structure (7mGpppN, where N is any nucleotide). Eukaryotic translation initiation factor (eIF) 4E of 25 kDa directly binds to the Cap and to a platform protein of 200 kDa, eIF4G. eIF4G interacts with the multi–subunit complex eIF3 (12 subunits; more than 500 kDa) and brings together the mRNA and the 43S pre–initiation complex formed by eIF3, the ternary complex (eIF2–Met–tRNAMet–GTP), the 40S ribosomal subunit and other initiation factors [1]. eIF4G also recruits the RNA helicase eIF4A which unwinds secondary structures in the 5' untranslated region (5'UTR) of the mRNA during the scanning towards the initiation codon, and the poly(A) binding protein (PABP) allowing the mRNA circularication for efficient translation re–initiation. Translation is probably the most controlled event in protein synthesis and an important regulatory mechanism takes place during Cap recognition and the mRNA recruitment steps [2].
The Cap–binding protein eIF4E has a highly conserved amino acid sequence in all organisms allowing its direct contact with the Cap structure [3]. eIF4G interacts with eIF4E through an YXXXXLΦ motif (where X is any amino acid and Φ is a hydrophobic residue) and improves its union with the Cap, forming a stable eIF4F–mRNA complex [4, 5]. The interaction between Cap and the translational machinery is prevented by the binding of eIF4E to other cellular proteins through the same motif used for its interaction with eIF4G. By such means cells could modulate either their global translation levels, or specific mRNA recruitment [6]. During the last few years it became evident that through binding to specific proteins and the Cap of mRNAs, eIF4E participates in the nucleo–cytoplasmic transport, translational repression, and turnover of mRNA.
Multiple eIF4E family members have been identified in a wide range of organisms that include plants, flies, mammals, frogs, birds, nematodes, and fish. These members have been classified into three families: eIF4E–I, eIF4E–II and eIF4E–III [7]. Some eIF4E family members have altered its Cap–binding affinities or the interaction with eIF4G and other proteins, providing clues to their physiological roles. It has been suggested that each organism has at least one eIF4E that is ubiquitous and constitutively expressed to carry out general translation, and that the other family members are involved in specialiced functions [2]. In addition, other proteins that do not belong to the eIF4E family, but are able to bind the Cap and perform a particular function during the RNA metabolism have been described. Such is the case of the nuclear Cap–Binding Complex (CBC) which participates in protection of the newly synthesiced transcripts and their export to the cytoplasm, the De–Capping protein S (DcpS) involved in mRNA degradation, and the Argonaute protein (AGO) in animals which is part of small RNA regulatory pathways (for review see [2]). Therefore, the proteic components of a Cap–binding complex may vary, depending upon the cellular conditions, growth and developmental requirements.
In plants, three eIF4E family members have been reported: eIF4E (ortholog of the mammalian eIF4E–1; class eIF4E–I) eIF(iso)4E (plant–specific; class eIF4E–I), and nCBP (novel Cap binding protein; class eIF4E–II). The eIF(iso)4E protein interacts with a particular eIF(iso)4G forming the unique plant eIF(iso)4F complex [8]. eIF4F and eIF(iso)4F complexes show selectivity in the recognition of mono and dimethylated Cap structures, as well as in in vitro translation of 5'UTR structured mRNAs. In most plant species, eIF(iso)4E shows about 50% amino acid identity with eIF4E and the relative abundance of each protein varies depending on the developmental stage and the plant tissue. In maice, the eIF(iso)4E protein is present at higher levels than eIF4E in non–germinated seeds [9]. The corresponding transcript is efficiently translated upon imbibition to maintain constant and high levels during the first 24 h of germination, whereas eIF4E levels increase toward germination completion. In addition, each Class I Zea mays eIF4E family member displays selective translational activity on the pool of mRNAs stored in the quiescent embryonic axes [10, 11].
Based on the above antecedents, in this work we aimed to identify the components of Cap–binding complexes at two particular developmental stages in maice: dry non–germinated (0h) and 24–h–imbibed germinated (24h) embryonic axes. Cap–binding proteins purified through affinity chromatography from 0h and 24h embryonic axes were separated on polyacrylamide gels, silver stained and identified by liquid chromatography–mass spectrometry (LC–MS–MS). This approach revealed a differential composition in the Cap–binding complexes from the two developmental stages, suggesting new roles for proteins as potential partners of the eIF4E family members.
Results and Discussion
Cap–binding protein patterns in dry (0h) and 24–h–imbibed (24h) embryonic axes
Maize Cap–binding complexes were purified from dry "non–germinated" (0h) and 24–h–imbibed (24h) "germinated" embryonic axes through m7GTP–affinity chromatography. Equal amounts of total protein extracts were used for the m7GTP–purification (Fig. 1A). After recovery of the m7GTP–bound fractions, equal volumes of each eluted fraction (F1–F4) were separated on 10% or 15% denaturing polyacrylamide gels for protein resolution of 200–45 kDa (Fig. 1B, upper gel) and 45–15 kDa (Fig. 1B, lower gel), respectively. The silver stained protein patterns indicated several differences between 0h– and 24h– Cap–binding complexes. First, several proteins were preferentially detected in the m7GTP–eluted fractions from 0h and were absent or decreased in the 24h embryonic axes (Fig 1A, dotted bands). Second, although the Cap–binding proteins in these complexes previously identified as eIF4E and eIF(iso)4E by western blot [9] were observed in both, 0h– and 24h– embryonic axes, their distribution in each eluted fraction was different, being eIF4E more tightly bound to the m7GTP–Sepharose than eIF(iso)4E (Fig. 1B, fraction F4 from lower gel). On the other hand, eIF(iso)4E and its corresponding partner eIF(iso)4G were mostly present in fractions F1 and F2. Third, a doublet was present at the position of eIF(iso)4G, whereas by western blot usually only one band is detected for this protein (see Fig. (2) 3). These results indicated that the composition and most importantly the affinity of the complexes bound to Cap vary between non–germinated and germinated maice seeds.
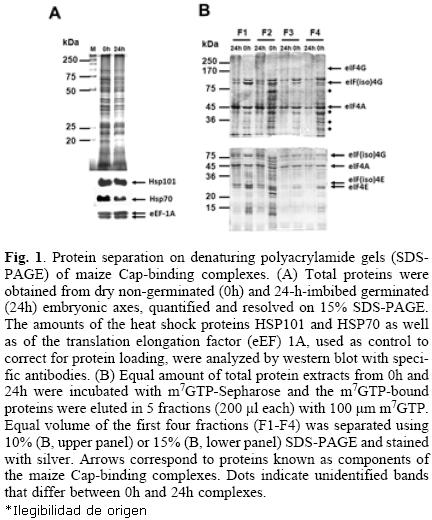

Protein identification in Cap–binding complexes from 0h and 24h embryonic axes
Several of the differential and conserved protein bands in the above purified Cap–binding complexes were selected for identification by mass spectrometry (Fig. 2; FT01–FT09). Proteins identified with the criteria of peptides with greater than 95% probability, and at least 2 identified peptides, are shown in Table 1. The complete information about the identified peptides is shwon in Tables 2 (0h) and 3 (24h). The analysis aimed to identify members of the translation initiation machinery and any associated proteins that may have potential translational regulatory roles during maice germination. It is important to notice, that there were silver–stained bands in the gel shown in Figure 2 not selected for identification by mass spectrometry in this study. These may include proteins present at both germination stages, or preferentially found at 24h. From the present analysis, eIF4E, eIF(iso)4E, and eIF(iso)4G proteins were identified in bands corresponding to their expected molecular weight. Noticeably, eIF4E was detected with only one peptide in the 0h– and with three peptides in the 24h–sample, whereas eIF4G was not identified in any of the processed bands. This is in agreement with previous findings in our lab, showing that the eIF(iso)4F is the most abundant Cap–binding complex in the dry maice seed [9].
The upper band of the doublet found at 88 kDa, in both 0h–and 24h–embryonic axes, rendered the identification of the 80 kDa subunit of the nuclear Cap–binding complex CBC. Other translation factors, such as eIF3c, eIF4A, and eEF1A were also identified in bands that correspond to their reported molecular weight. According to the literature, these proteins are usually found in Cap–binding complexes from other organisms [12, 13]. The role of eIF3c and eIF4A is at the level of translation initiation, whereas eEF1A is the elongation factor that carries incoming aminoacylated tRNA to the A site of the ribosome. Since eIF4E and eIF4G proteins remain bound to the mRNA to allow efficient re–initiation of translation on a circulariced mRNA, both initiation and elongation factors may be found within the Cap–binding complexes.
Recently, the protein degradation and synthesis machineries have been reported as complexes sharing several of their components [14]. In agreement, we found the 6A subunit of the 26S proteasome in the Cap–binding complex from 0h–em–bryonic axes. This protein is a component of the small subunit (19S) of the proteasome. According to the literature, several proteins from the eIF3 multisubunit complex may interact with proteins of the proteasome and are specifically targeted for degradation [14].
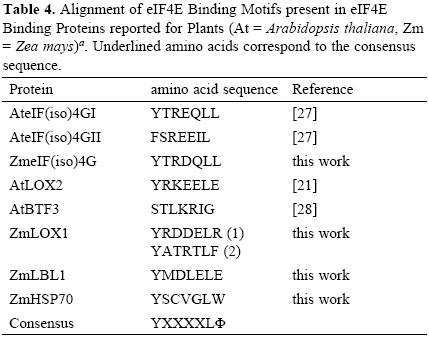
Interestingly, two heat shock proteins, HSP101 and HSP70, were identified as part of the Cap–binding complexes in the dry, but not in the 24–h–imbibed embryonic axes. These proteins were detected at similar levels in the total protein extracts from both samples (Fig. 1A), suggesting that a possible change of their interaction with components of the Cap–binding complex, instead of degradation, is taking place during maice germination. The chaperone HSP70 has been previously identified in Cap–binding complexes in Drosophila melanogaster [12]. This chaperone is usually associated with the polypeptide chains nascent from the ribosomal large subunit to assure correct folding early in protein synthesis. Its presence in the Cap–binding complex from 0h–embryonic axes may be indirect through the association with the nascent polypeptide chain or even more likely due to its requirement in assisting the folding of translation initiation factors. However, a closer look on the amino acid sequence of maize HSP70 revealed the YXXXXLΦ conserved motif of eIF4E interacting proteins (Table (3) 4). This suggests that maice HSP70 may directly bind to eIF4E and regulate its function in mRNA recruitment. Supporting this, proteins from the HSP70 family have been shown to regulate translation and stability of mRNAs with AU–rich element (ARE) in their 3'UTRs [15]. However, the specific binding of HSP70 to any of the known Cap–binding translation factors must be further confirmed.
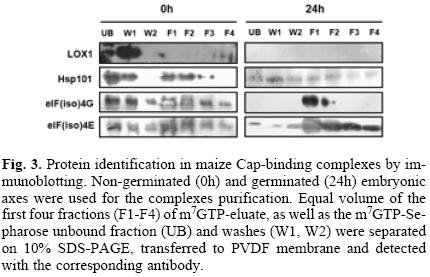
The other heat shock protein, HSP101, has been reported as a chaperone involved in disaggregation of large protein complexes and its expression is greatly induced under heat shock. Maize HSP101 is accumulated in the dry seed and is required to achieve thermotolerance in young germinating seedlings [16]. During the first 24 h after seed imbibition, its protein level remains unchanged, but after 72h of seed imbibition HSP101 is almost undetectable under normal temperature growth conditions (25–32 °C). Wheat HSP101 has been reported as trans–lational regulator for specific mRNAs harboring particular sequences in their UTRs [17]. In maice, a null mutant for this chaperone displays accelerated root growth during germination under normal temperature conditions, but under heat shock, the successful seedling establishment is impaired [16]. Therefore, the presence of HSP101 in the 0h Cap–binding complexes may account for either protein disaggregation of translation factors needed to guarantee translation initiation of growth regulators as soon as the seed is imbibed, or for translation regulation of particular mRNAs during early germination.
Surprisingly, several proteins belonging to carbohydrate metabolic pathways were also detected in the 0 h–, but not in the 24 h– Cap–binding complexes. These included the 3–phosphoglycerate kinase, fructose biphosphate aldolase, glyceralde–hyde–3–phosphate dehydrogenase, malate dehydrogenase and alcohol dehydrogenase1. According to the number of identified peptides for each of these proteins (Table 1), they appear as abundant in the Cap–binding complexes. One possibility might be that their synthesis is stopped during the desiccation process and hence they may co–purify as nascent polypeptides within translation complexes. On the other hand, they may form part of aggregates together with translation factors. Although translational functions for these proteins have not been described yet, in a recent work reporting the yeast translasome several metabolic encymes co–purified with the eIF3 translation complex [18]. In addition, the glyceraldehyde 3–phosphate dehydrogenase has been associated to nuclear RNA export [19] suggesting that metabolic encymes may have additional functions within protein synthesis. The presence of actin as part of the Cap–binding complexes is not unexpected, since translation complexes are bound to the cytosqueleton of the cell and this protein has been found as critical for normal protein synthesis in mammalian cells [20].
Maize Lipoxygenase 1 (LOX1) and the trans–acting siRNA (tasiRNA) biogenesis–related protein Leafbladeless 1 (LBL1) are not abundant proteins in the maice seed and were as well detected in the Cap–binding complexes of dry seeds (Table 1; FT02 and FT03). A member from the Lipoxygenase family, the Arabidopsis thaliana LOX2, was previously reported as eIF(iso)4E binding protein that contains a putative conserved YXXXXLΦ motif [21], whereas LBL1 has been found as meristem specific protein involved in small RNA mediated gene silencing and abaxial/adaxial leaf fate definition [22]. Therefore, we searched the sequence of maice LOX1 and LBL1 for the YXXXXLΦ motif to find whether these are also potential eIF4E or eIF(iso)4E binding proteins. The analysis showed that both proteins presented the conserved motif (Table 4). Since eIF(iso)4G also uses this amino acid sequence to bind eIF(iso)4E and integrate a functional translation initiation complex on the mRNA, the presence of potential eIF(iso)4E binding proteins in the 0h Cap–binding complexes indicates that eIF(iso)4E may be a part of ribonucleoprotein particles not involved in translation initiation at this developmental stage.
HSP101 and LOX1 presence in the Cap–binding complexes from dry and 24–h–imbibed maize seeds
To test whether some of the newly identified proteins in Cap–binding complexes are indeed specifically eluted from the m7GTP–chromatography, western blot with available antibodies was performed with extended washes before the specific elution with the m7GTP ligand (Fig. 3). While the LOX1 protein was mostly detected in the first washes (W1) of the column, a small quantity was specifically retained and eluted with the ligand in the last fractions (F2–F4) from dry (0h) embryonic axes. This protein was not detected in the 24–h–imbibed axes neither in total protein extracts nor in the Cap–bound fractions, indicating that it is probably particularly expressed during seed maturation. A significant amount of HSP101 was also specifically retained in the column, but eluted in the first three fractions upon the ligand addition (F1–F3), when non–germinated embryonic axes were used. Interestingly, the protein was also detected in the 24–h–imbibed embryonic axes elution fractions, although to a lesser levels. This could correlate to previous data in wheat, where HSP101 was reported to bind to eIF4G and eIF3, but not to eIF4E proteins [23]. On the other hand, the elution pattern of LOX1 was similar to that observed for eIF(iso)4E, indicating a possible direct binding between these two proteins. These results support the notion that eIF(iso)4E, which is the major Cap–binding translation initiation factor in the dry non–germinated maice seed, forms different multiproteic complexes to regulate selective mRNA translation upon the germination onset.
Conclusions
The analysis of Cap–binding complexes at two different germination stages in the maice seeds indicated differential composition that may correlate with the translational requirements and regulatory mechanisms operating to achieve the appropriate protein synthesis patterns at each developmental stage. New components of the Cap–binding complexes in non–germinated seeds include the chaperones HSP101 and HSP70 as well as the lipoxygenase LOX1 and leafbladeless LBL1. HSP70, LOX1 and LBL1 are candidates to interact with members of the eIF4E family through an YXXXXLΦ motif.
Experimental
Material and Methods Plant material
Maize (Zea mays L) seeds of a Mexican land race Tuxpeño, var. Chalqueño, were used for all experiments. Seeds were germinated by water imbibition on moisturiced cotton, in the dark, at 25 °C. Embryonic axes were manually excised from either dry (0h) or 24–h–imbibed (24h) seeds.
Cap–binding complexes purification
Cap–binding complexes were purified by m7GTP–Sepharose affinity chromatography as previously reported [9], with some modifications. Briefly, 2.5 g of axes were macerated in liquid nitrogen and suspended in 25 mL Buffer "A" consisting of: 20 mM HEPES, pH 7.6; 100 mM KCl; 0.2 mM EDTA; 10 % glycerol; 1 % Triton X–100; 0.5 mM DTT; and CompleteTM, EDTA free protease inhibitors (CompleteTM, Roche Molecular Diagnostics, Pleasanton, CA, USA). The extract was clarified by 30 min centrifugation at 15,000 rpm and 4 °C in a Sorvall J–20 rotor. The supernatant was filtered through eight layers of cheesecloth and the protein amount was quantified. Approximately, 20 mg of total protein was incubated with 0.5 mL of m7GTP–Sepharose (GE Healthcare Bio–Sciences AB, Uppsala, Sweden) for one h at 4 °C. The slurry was poured onto a 10 mL Column (Bio–Rad Laboratories, Inc. Hercules, CA) and the resin was washed with 5 mL of Buffer "A", followed by 5 mL of Buffer "A" including 0.1 mM GTP. The bound proteins were eluted with 1 mL of Buffer "A" containing 100 uM m7GTP (Sigma–Aldrich Co., Saint Louis MO, USA) in 5 fractions of 200 uL each. To assess more–specific binding, washes with Buffer "A" were extended to 10 mL (20 bed volumes) divided in 5 mL each.
Electrophoresis and protein staining
Proteins from the purified Cap–binding complexes were resolved on either 10% or 15% (w/v) denaturing polyacrylamide (SDS–PAGE) gels. The silver staining procedure used was compatible with mass spectrometry analysis [24].
Mass spectrometry
The in–gel digest and mass spectrometry experiments were performed by the Proteomics platform of the Eastern Quebec Genomics Center, Quebec, Canada. Tryptic digestion was performed on a MassPrep liquid handling robot (Waters, Milford, USA) according to the manufacturer's specifications and to the protocol of [24] with the modifications suggested by [25]. Briefly, proteins were reduced with 10 mM DTT and alkylated with 55 mM iodoacetamide. Trypsin digestion was performed using 105 mM of modified porcine trypsin (Sequencing grade, Promega, Madison, WI, USA) at 58°C for 1h. Digestion products were extracted using 1% formic acid, 2% acetonitrile followed by 1% formic acid, 50% acetonitrile. The recovered extracts were pooled, vacuum centrifuge dried and then resus–pended into 8 uL of 0.1% formic acid and 4 uL were analyced by mass spectrometry.
Peptide samples were separated by online reversed–phase (RP) nanoscale Capillary liquid chromatography (nanoLC) and analyced by electrospray mass spectrometry (ES MS/MS). The experiments were performed with a Thermo Surveyor MS pump connected to a LTQ linear ion trap mass spectrometer (ThermoFisher, San Jose, CA, USA) equipped with a nanoelec–trospray ion source (ThermoFisher). Peptide separation took place on a PicoFrit column BioBasic C18, 10 cm x 0.075 mm internal diameter (New Objective, Woburn, MA, USA), with a linear gradient from 2–50% solvent B (acetonitrile, 0.1% formic acid) in 30 min, at 200 mL/min (obtained by flow–splitting). Mass spectra were acquired using a data dependent acquisition mode using Xcalibur software version 2.0. Each full scan mass spectrum (400 to 2000 m/c) was followed by collision–induced dissociation of the seven most intense ions. The dynamic exclusion (30 sec exclusion duration) function was enabled, and the relative collisional fragmentation energy was set to 35%.
Protein Identification
All MS/MS samples were analyced using Mascot (Matrix Science, London, UK; version 2.2.0). Mascot was set up to search the ncbi_Zea_mays_20071004 database (10,023 entries) assuming the digestion encyme non–specific. Mascot was searched with a fragment ion mass tolerance of 0.50 Da and a parent ion tolerance of 2.0 Da. Iodoacetamide derivative of cysteine was specified as a fixed modification and oxidation of methionine was specified as a variable modification. Scaffold (version 3.0, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Peptide Prophet algorithm [26] and contained at least 2 identified peptides. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
Immunoblotting
For immunodetection, proteins were blotted onto a polyvinylidene fluoride (PVDF) membrane (Millipore Corp., Billerica, MA, USA), which was blocked with 5% (w/v) milk and incubated with the primary antibody for 2 h at room temperature. After several washes in Phosphate Saline Buffer (PBS), the membrane was incubated for 1 h with the appropriate secondary antibody at a 1:5000 dilution. Detection was performed with Immobilon Western Chemiluminescent HRP Substrate (Millipore Corp.). Primary antisera dilutions were as follows: antibodies against wheat eIF(iso)4E and eIF(iso)4G were kindly donated by Karen S. Browning, University of Texas, Austin, USA and were used at 1:5000 dilution; antibody against maice Hsp101 was kindly donated by Jorge Nieto–Sotelo, Instituto de Biotecnología, UNAM, Cuernavaca, Mexico and used at 1:1000 dilution; antibody against bean LOX2 was kindly donated by Helena Porta, Instituto de Biotecnología, UNAM, Cuernavaca, Mexico and used at 1:100 dilution.
Acknowledgements
The authors acknowledge the financial support from Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica, México, IN204309 and Consejo Nacional de Ciencia y Tecnología, México, 81708. We wish to thank Dr. Karen S. Browning (University at Austin, Texas), Dr. Jorge Nieto–Sotelo and Dr. Helena Porta (Instituto de Biotecnología, Universidad Nacional Autónoma de México) for antibody donations.
References
1. Jackson, R. J.; Hellen, C. U.; Pestova, T. V. Nat. Rev. Mol. Cell Biol. 2010, 11, 113–127. [ Links ]
2. Rhoads, R. E. J Biol Chem. 2009, 284, 16711–16715. [ Links ]
3. Goodfellow, I. G.; Roberts, L. O. Int. J. Biochem. Cell Biol. 2008, 40, 2675–2680. [ Links ]
4. Mayberry, L. K.; Allen, M. L.; Dennis, M. D.; Browning, K. S. Plant Physiol. 2009, 150, 1844–1854. [ Links ]
5. Kaye, N. M.; Emmett, K. J., Merrick; W. C.; Jankowsky, E. J Biol Chem. 2009, 284, 17742–17750. [ Links ]
6. Sonenberg, N.; Hinnebusch, A. G. Mol Cell. 2007, 28, 721–729. [ Links ]
7. Joshi, B., Lee, K.; Maeder, D. L.; Jagus, R. BMC Evol. Biol. 2005, 5, 48–68. [ Links ]
8. Browning, K. S. Biochemical Society Transactions. 2004, 32, 589-591. [ Links ]
9. Dinkova, T. D.; Sanchec de Jimenec, E. Physiol. Plant. 1999, 107, 419–425. [ Links ]
10. Dinkova, T. D.; Aguilar, R.; Sanchec de Jimenec, E. In The Biology of Seeds: Recent Research Advances (Nicolas, G., Bradford, K. J., Come, D. and Pritchard, H. W., eds.). 2003, pp. 181–189, CAB International. [ Links ]
11. Dinkova, T. D.; Marquec–Velacquec, N. A.; Aguilar, R.; Lacaro–Mixteco, P.; Sanchec de Jimenec, E. Seed Science Research. 2011, 21, 85–93. [ Links ]
12. Hernandec, G.; Altmann, M.; Sierra, J. M.; Urlaub, H.; Diec del Corral; R., Schwartc, P.; Rivera–Pomar, R. Mech. Dev. 2005, 122, 529–543. [ Links ]
13. Fierro–Monti, I.; Mohammed, S.; Matthiesen, R.; Santoro, R.; Burns, J. S.; Williams, D. J.; Proud, C. G.; Kassem, M.; Jensen, O. N.; Roepstorff, P. J Proteome Res. 2006, 5, 1367–1378. [ Links ]
14. Cabrera, R.; Kleifeld, O.; Scheliga, J. S.; Glickman, M. H.; Chang, E. C.; Wolf, D. A. Mol Cell. 2009, 36, 141–152. [ Links ]
15. Laroia, G.; Cuesta, R.; Schneider, R. J. Science 1999, 284, 499–502. [ Links ]
16. Nieto–Sotelo, J.; Martinec, L. M.; Ponce, G.; Cassab, G. I.; Alagon, A.; Ribaut, J. M.; Yang, R. Plant Cell. 2002, 14, 1621–1633. [ Links ]
17. Ling, J.; Wells, D. R.; Tanguay, R. L.; Dickey, L. F.; Thompson, W. F.; Gallie, D. R. Plant Cell. 2000, 12, 1213–1227. [ Links ]
18. Sha, Z.; Brill, L.M.; Cabrera, R.; Kleifeld, O.; Scheliga, J.S.; Glickman, M.H. Chang, E.C.; Wolf, D.A. Mol Cell. 2009, 36, 141–152. [ Links ]
19. Sirover, M.A. J Cell Biochem. 2005, 95, 45–52. [ Links ]
20. Stapulionis, R.; Kolli, S.; Deutscher, M.P. J. Biol. Chem. 1997, 272, 24980–24986. [ Links ]
21. Freire, M. A.; Tourneur, C.; Granier, F.; Camonis, J.; El Amrani, A.; Browning, K. S.; Robaglia, C. Plant Mol. Biol. 2000, 44, 129–140. [ Links ]
22. Timmermans, M.C.; Schultes, N.P.; Jankovsky, J.P.; Nelson, T. Development. 1998, 125, 2813–2823. [ Links ]
23. Gallie, D. R. Nucleic Acids Res. 2002, 30, 3401–3411. [ Links ]
24. Shevchenko, A.; Wilm, M.; Vorm, O.; Mann, M. Analytical Chemistry. 1996, 68, 850–858. [ Links ]
25. Havlis, J.; Thomas, H.; Sebela, M.; Shevchenko, A. Analytical Chemistry. 2003, 75, 1300–1306. [ Links ]
26. Keller, A.; Nesvichskii, A.; Kolker, E.; Aebersold, R. Analytical Chemistry. 2002, 74, 5383–5392. [ Links ]
27. Lellis, A. D.; Allen, M. L.; Aertker, A. W.; Tran, J. K.; Hillis, D. M.; Harbin, C. R.; Caldwell, C.; Gallie, D. R.; Browning, K. S. Plant Mol. Biol. 2010, 64, 249–263. [ Links ]
28. Freire, M. A. Gene 2005, 345, 271–277. [ Links ]
Note
Dedicated to Dr. Estela Sánchez de Jiménez for her invaluable contributions to plant biochemistry.














