Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Similares em
SciELO
Similares em
SciELO
Compartilhar
Tropical and subtropical agroecosystems
versão On-line ISSN 1870-0462
Trop. subtrop. agroecosyt vol.13 no.1 Mérida Jan. 2011
Artículos de investigación
Genetic diversity and symbiotic efficiency of legume nodulating bacteria from different land use systems in Taita Taveta, Kenya
Diversidad genética y eficiencia simbiótica de bacterias noduladoras de leguminosas obtenidas de sistemas con diferentes usos de suelo en Taita Taveta, Kenia
S. N. Mwangi1*, N. K. Karanja2, H. Boga1, J. H. P. Kahindi3, A. Muigai1, D. Odee4 and G. M. Mwenda1
1 Jomo Kenyatta University of Agriculture and Technology, P.O. Box 62000-00200, Nairobi * Corresponding author Email: mwangingare@gmail.com
2 University of Nairobi, P.O Box 30197-00100, Nairobi.
3 United States International University, P.O Box 14634-00800, Nairobi.
4 Kenya Forestry Research Institute, P. O. Box 20144 Nairobi, Kenya.
Submitted July 27, 2010
Accepted September 22, 2010
Revised received September 30, 2010
Abstract
Populations of Legume Nodulating Bacteria (LNB) were assessed under glasshouse conditions from soils collected in Taita Taveta district, Kenya from various landuse systems. The populations were estimated by the most-probable-number (MPN) plant infection technique using Macroptilium atropurpureum (DC.) Urban (siratro) as the trap plant. The LNB populations varied from 1.1 x 10 to 6.1 x 106 cells g-1 of soil. There was apparent landuse effect on abundance of LNB with maize-bean cropping system and Shrubland giving high population estimates. Two thousand isolates of LNB were obtained from the nodules of siratro trap plant. These isolates were characterized on yeast extract mannitol mineral salts agar (YEMA) media containing bromothymol blue and two distinct rhizobia growth rate types were identified: fast growers (acid-producing) at 78.6% while slow growers (alkali-producing) comprised 21.4%. Symbiotic effectiveness of a selected number of the isolates ranged from 6.7% to 96.4% and no clear influence of landuse was observed. RFLP of amplified 16S rRNA genes of isolates with HaeIII and TaqI grouped the isolates into seven ribotypes and partial sequencing of 16S rRNA genes of isolates representative of the ribotypes further grouped the isolates into six genera namely; Sinorhizobium, Bradyrhizobium, Herbaspirillum, Agrobacterium, Rhizobium and Burkholderia. Landuse type was found to significantly influence the diversity of LNB (PO.05). The highest LNB richness of five genera was found in indigenous forest soils. While fallow/shrubland and maize based system had a total richness of four genera. Each of the remaining landuses had LNB total richness of two.
Key words: Diversity; indigenous LNB; landuse systems; most-probable-number (MPN); Symbiotic effectiveness; RFLP, Macroptilium atropurpureum (Siratro).
INTRODUCTION
Symbiotic association between legumes and Leguminosae Nodulating Bacteria (LNB) is an important biocatalytic link for the flow of nitrogen between available nitrogen reservoir, the atmosphere and the living world (Paul and Clark, 1989). Exploitation of the legume-rhizobia symbiosis in agricultural systems requires knowledge of LNB available in different agro-ecological zones as foreign strains introduced as inoculants often fail to adapt well (Cheng et al., 2009). There are several reports on studies on natural nodulation of agriculturally important pasture and grain legumes in cropping systems of Kenya (McDonald, 1935; Bumpus, 1957; Morrison, 1966; Souza, 1969). Most of these legumes have been reported to nodulate with varying levels of nodulation intensity- from poor or erratic to very profuse nodulation. However, these earlier studies did not quantify the abundance and genetic characteristics of indigenous LNB populations. More recently, Odee et al. (1995) surveyed natural nodulation and determined the abundance of indigenous populations in a wide spectrum of agro-ecological zones mostly from indigenous woodlands. The isolates from these systems showed a wide range of phenotypic and genetic diversity, which also indicated that most of the described genera were present (Odee et al., 1997, 2002). Other studies by Anyango et al. (1995, 2005) also pointed towards considerable diversity of LNB in Kenya. A common feature of these studies is that they did not investigate the influence of different landuse systems on the diversity of LNB. Landuse has been shown to influence diversity and abundance of LNB although relationships are not clear (Martinez-Romero and Caballero-Mellado, 1996; Ngokota et al., 2008; Zawedde et al., 2009). In this study, we investigated abundance and diversity of LNB occurring in various agro-ecological zones including both intensely cultivated agricultural systems and undisturbed indigenous forests in TaitaTaveta district, Kenya. Symbiotic efficiencies of LNB isolates from the area were also assessed.
MATERIAL AND METHODS
Description of the study site and selection of sampling points
The study was conducted in Taita Taveta District, located in South-eastern Kenya, latitude 03° 20' S longitude 38° 15' E and at an altitude of 2228 m above sea level. The area receives an average annual rainfall of 1500 mm in the highlands and 250 mm in the lowlands and the mean monthly temperatures range from 17.4° C to 34.5° C. The soils are primarily sandy loam with high infiltration rates, low pH, low water holding capacity, and low nutrient contents due to excessive leaching. With the aid of GPS, the study area was stratified based on five land uses identified as being dominant in the area. These land use systems were: Maize-based fanning (Zea mays), Shrubland (mainly Lantana cámara), indigenous forest, agroforestry and planted forest. Points, 200 m apart were marked with the aid of GPS to fall within all the land use systems. From these, five points were picked randomly from each land use.
Soil Sampling
At each sampling point, two vertically crossing lines and two concentric circles of radius 3 m and 6 m were drawn. An auger of 7 cm diameter was used to take four cores of soil from the 0-20 cm depth in the small circle and eight in the outer circle. The 12 subsamples were homogeneously mixed to constitute a composite sample from which 500 g soil was taken, placed in a plastic bag, and double sealed and then kept under shade. The soil auger was sterilized with ethanol between sampling points to avoid cross contamination. The soil samples were transported to the laboratory where they were kept at room temperature before isolation of rhizobia. Soil physical-chemical analysis at the sampling points was also done as already reported by Karanja et al., 2009.
Seed pre-treatment and germination
Healthy siratro (Macroptilium atropurpureum (DC.) Urban) seeds of uniform size were placed in a sterile Erlenmeyer flask and covered with a sterile Petri dish. Concentrated sulphuric acid was added to coat the seeds. Sterilization and scarification was allowed to proceed for 10 min before draining off excess acid. Sufficient volume of sterile water was added to first rinse the acid. The seeds were rinsed in another five changes of sterile water and then left overnight in a refrigerator at 4°C to imbibe. The pre-treated seeds were further rinsed in two changes of sterile water then germinated on 0.75% (w/v) agar plates.
Enumeration of LNB
Indigenous LNB populations were determined using the plant infection technique (Somasegaran and Hoben, 1994). Each composite soil sample was mixed thoroughly and quartered. Soil inocula were prepared by suspending 10 g of soil sample in 90 ml of sterile water in a 160-ml dilution bottle and shaken for 20 min in a wrist-action shaker at room temperature (~25°C). 1 ml of each suspension was aseptically pipetted into 9 ml of sterile water diluent in McCartney bottle and shaken for 2 min. The resulting suspension was serially-diluted tenfold from 10-1 to 10" 6 with four replications at each dilution level. Aliquots of 1 ml were used to inoculate 3-5 day old siratro seedlings previously pre-treated, germinated and aseptically transferred to sterile plastic pouches containing N-free nutrient solution (Broughton and Dilworth, 1971) with two plants per pouch. The pouches were supported in improvised racks. The plants were grown for 28 days in a glasshouse at temperature 30/18 °C (day/night) and natural light of ca. 12 h photoperiod. The number of nodulated plants at each dilution was recorded and used as an ordered code (from low to high dilution) and used to estimate the most-probable-number (MPN). The computer program, Most Probable Number (MPN) by Bennet et al. (1990) was used to calculate the populations.
Isolation of Rhizobia from Siratro root nodules and morphological characterization
Nodules were surface sterilized in 1 % NaOCl for 6 min, rinsed in several changes of sterile water, and then crushed with a flame-sterilized blunt-tipped pair of forceps. A loopful of the crushed nodule was then streaked across the surface of Petri dish containing yeast extract mannitol mineral salts agar (YEMA) (Vincent, 1970). Some nodules had dual or multiple nodule occupancy; not all nodules produced isolates. Typical well-isolated colonies were re-isolated and characterized on YEMA containing 25 mg kg-1 (w/v) bromothymol blue (BTB) as a pH reaction indicator. In addition, the growth of the isolates was characterized by the rate of colony emergence on YEMA/BTB media incubated at 28°C. Fast- and slow-growing LNB were described as emerging after three to five days and seven to ten days following inoculation, respectively. All isolates were stored in 16% glycerol yeast mannitol broth (YMB) at -70°C.
Symbiotic efficiency test
Nitrogen fixation potential was determined as described by Somasegaran and Hoben (1994). Siratro planted in modified Leonard jar assembly (Leonard, 1943) with vermiculate as growth media and nitrogen free nutrient solution (Broughton and Dilworth, 1971), were inoculated with isolate to be tested. Non-inoculated nitrogen-free and nitrogen-supplemented plants were used as controls. The experiment was conducted in the greenhouse for eight weeks after which biomass and nitrogen fixation effectiveness were determined as described by Gibson (1987): Shoot dry weight (SDW) and the ratios of inoculated plants/ non-inoculated nitrogen supplemented control.
PCR amplification of bacterial 16S rRNA gene
The total genomic DNA was extracted by alkaline lyses method (Sambrook et al, 1989). The extracted total DNA from each sample was used as a template for amplification of the 16S rRNA gene using universal primers 8F (5'- AGAGTTTGATCATGGCTCAG-3') and 1492R (5'-GGTTACCTTGTTACGACTT-3'). Amplification was performed using a model 9700 Fast Thermal Cycler from Applied Biosystems. Amplification was carried out in a 30 mixture containing 3
mixture containing 3  l of 1 Ox PCR buffer, 4
l of 1 Ox PCR buffer, 4 l of 2.5 mM dNTPs, 2.5
l of 2.5 mM dNTPs, 2.5  l of 27F forward primer (5 pmol), 2.5
l of 27F forward primer (5 pmol), 2.5 l of 1492R reverse primer (5 pmol), 0.4
l of 1492R reverse primer (5 pmol), 0.4  l of 5U/ul Taq polymerase, 1.5
l of 5U/ul Taq polymerase, 1.5  l template DNA and 16.6
l template DNA and 16.6  l of PCR grade water. Reaction mixtures were subjected to the following temperature cycling profiles: Initial denaturation at 94° C for 5 minutes, 30 cycles of denaturation at 94° C for 45 seconds, primer annealing at 55° C for 50 seconds, chain extension at 72° C for 90 seconds, and a final extension at 72°C for 8 minutes. Amplification products (5
l of PCR grade water. Reaction mixtures were subjected to the following temperature cycling profiles: Initial denaturation at 94° C for 5 minutes, 30 cycles of denaturation at 94° C for 45 seconds, primer annealing at 55° C for 50 seconds, chain extension at 72° C for 90 seconds, and a final extension at 72°C for 8 minutes. Amplification products (5 l) of each DNA sample was loaded on agarose gel (1 %) containing ethidium bromide and run in IX TAE buffer at 80 volts for 1 hour. Gel documentation was done using the Gel Logic 200 Imaging System (Sambrook et al, 1989).
l) of each DNA sample was loaded on agarose gel (1 %) containing ethidium bromide and run in IX TAE buffer at 80 volts for 1 hour. Gel documentation was done using the Gel Logic 200 Imaging System (Sambrook et al, 1989).
RFLP analysis of 16S rRNA gene
PCR products were digested with HaeIII and TaqI restriction enzymes (Boehringer Mannheim, meylan, France) according to manufacturer's instructions. RFLPs were analyzed in 1.8 % agarose gel in TAE buffer at 60 volts for 1.5 hours. Gel visualization was done using the Gel Logic 200 Imaging System (Sambrook et al, 1989).
PCR amplicons purification, Sequencing and phylogenetic analysis
The PCR amplicons of the selected isolates were purified using quickClean PCR purification kit (GenScript Corporation, 120 Centennial Ave, Piscataway, NJ 08854) according to the manufacturer's instructions. Partial sequencing of Purified PCR products was done at International Livestock Research Institute (ILRI) commercial lab (Segolilab) using 8F and 1492R primers as described byHureke/a/., (1997).
Sequence results were edited using Chromas software. The 16S rRNA gene sequences were compared to sequences in the public database using Basic Local Alignment Search Tool (BLAST) on the National Center for Biotechnology Information (NCBI) website in order to determine similarity to sequences in the Genebank database (Shayne et al, 2003). The 16S rRNA gene sequences determined in this study were aligned with highly similar sequences from Genbank with Clustal W. The evolutionary history was inferred using the Neighbor-Joining method (Saitou & Nei, 1987). Computation of evolutionary distances were done using the Maximum Composite Likelihood method (Tamura et al, 2004) and Phylogenic analyses were conducted in MEGA4 (Tamura et al, 2007). Bootstrap for 500 replicates was performed to attach confidence estimates for the free topologies (Felsenstein, 1985).
RESULTS AND DISCUSSION
Indigenous LNB populations in Taita soils
The populations of indigenous LNB nodulating siratro in soils collected from sampling points across some of land use systems are presented in Table 1. All soils induced nodulation. Population of LNB varied from one landuse system to the other. The highest population size detectable by this assay was 6.1 x 106 cells g-1 soil in the Shrubland while the lowest was 1.1 x 10 cells g-1 soil in soils from the agroforestry systems.
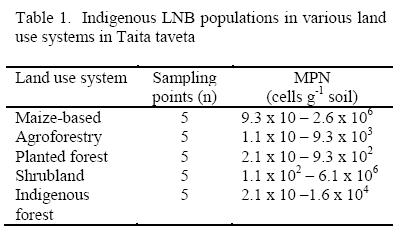
The population sizes determined in these sampling points were within the ranges reported for LNB associated with native woody legumes (mainly Acacia spp.) occurring in diverse ecological regions of Kenya (Odee et al, 1995). There were large variations within landuse systems perhaps due to differences in soil fertility and land use intensity. Shrubland and maize-based systems had some of the highest LNB populations. This is probability due to the presence of various leguminous plant species in Shrubland and the presence of the common bean (Phaseolus vulgaris) in the maize-based system. It has been demonstrated that indigenous common bean nodulating LNB occurring in acid soils (pH ≤ 4.5) of Kenya have a broad host range that include siratro (Anyango et al. 1995).
Characterization of LNB by growth rate
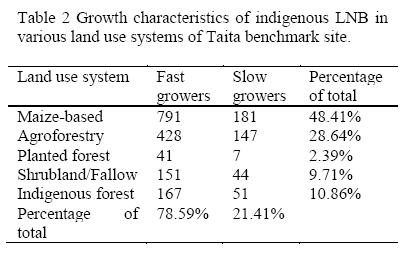
A total of 2008 pure isolates were recovered from root nodules of siratro MPN plants. Dual or multiple nodule occupants which was common is a common phenomenon in nodulated legumes growing in tropical soils (Odee et al, 1997). The pure isolates fell into two major growth rate types: fast growers (acid-producing) and slow growers (alkali-producing). More fast-growing types (78.59%) were isolated than slow-growing types (21.41%) as shown on Table 2. Most of the isolates (48.41%) were from the maize-beans landuse system probably also due to the presence of the common bean which is known to be nodulated by a wide range of rhizobia (Sawada et al., 2003). Fast growers were acid-producing and slow growers were alkali-producing on yeast extract mannitol mineral salts agar (YEMA) media incubated at 28°C.
Characterization of LNB isolates by morphology
Morphological characterization of the 2008 LNB isolates gave rise to twenty eight (28) morphtypes (Table 3). Morphtype 2 was the most dominant (29.03%) followed by morphotype 1 (19.77%). Almost half of the isolates fell into the two morphotypes. Morphotype 27 was the least abundant with only 2 isolates (0.10%) falling into this morphological group.
Observed morphological characteristics as shown in Table 3 indicate that there was high diversity within the isolates trapped with siratro from Taita soils. Diversity was high even between the two groups earlier identified based on growth rates. Previous studies have shown that cultural and morphological characteristics are not sufficient to fully characterize tropical LNB populations (Zhang et al. 1991; Odee et al. 1997; Bala et al. 2004). However, these characteristics, together with consideration for landuse representativeness, were used to scale down isolates from a large number of 2008 to a 100. These 100 isolates were assessed for their symbiotic efficiency in association with siratro and then further characterized by sequencing of 16SrRNA genes (Odee et al., 2002) to describe higher level (genera and species) taxa.
Symbiotic efficiencies of LNB
Of the hundred isolates selected to be tested for symbiotic efficiencies (SE) with siratro, 72 were ineffective and 28 re-infected which conforms to observations reported by (Anyango et al., 1995; Khbaya et al, 1998; Odee et al, 2002). There were significant differences in symbiotic efficiencies among the isolates. SE ranged from a low of 6.7% in isolate 767, which was almost similar to 'SE' of the negative control (7.14%), to a high of 95.4% in the case of isolate 6 (Figure 1). Approximately 18% of isolates had SE values of above 50%. A previous study with almost similar number of isolates (32) found 9% of isolates to have SE of above 50% (Laranjo et al, 2001). 5 isolates with high SE were from soils under varied landuse systems.
Molecular characteristics of isolates
Genomic DNA was successfully extracted from the 28 isolates that had effectively re-nodulated siratro in the previous symbiotic efficiency testing step. Genomic DNA was also extracted from 3 isolates picked randomly from those that failed to re-nodulate siratro. PCR of the 16S rRNA gene loci from each of the strains produced a single band of 1·5kb which corresponds to the expected size reported previously (Terefework et al, 1998). Restriction of PCR products with Haelll gave rise to seven (7) ribotypes (Figure 2) while restriction with TaqI gave minimal resolution (data not shown).
Some ribotypes were displayed by more isolates than others. An example is the ribotype represented in lanes 1, 3, 5, 7, 8, 11, 12, 15, 16, and 17, among others. Less common ribotypes included the one exhibited by isolate ran on lane 6. 7 isolates, one to represent each ribotype, were picked and had their 16 S rRNA genes sequenced. After editing, aligned sequences of representatives of all ribotypes and those of formally described close relatives from NCBI, were phylogenetically analyzed using MEGA 4.0. Two of the ribotypes clustered with Bradyrhizobium, one with Sinorhizobium, one with Rhizobium, another one with Agrobacterium and the other two with Burkholderia vietnamiensis and Herbaspirilium lineage (Figure 3).
Phylogenetic analysis revealed that isolates fell roughly into six genera and their distribution differed with landuse as shown in Table 4.
Among the genera were Sinorhizobium, Burkholderia, and Herb aspir ilium. These 3 genera, unlike Agrobacterium, Rhizobium and Bradyrhizobium, have not been commonly described from Kenyan soils. Herbaspirillum species are a group of diazotrophs initially commonly found in association with cereals (Baldani et al., 1986) but have since been continually found in association with legumes. Olivares et al. (1996) described the isolation of H. seropedicae not only from gramineae but also from roots of a legume species (Cajanus cajari). Valverde et al. (2003) has since confirmed Herbaspirillum species can indeed fix nitrogen in association with legumes. Herbaspirillum sp. was found in only soils under indigenous forest which is indicative of its narrow host range with legumes.
An isolate closely related to Sinorhizobium fredii was also identified. Sinorhizobium fredii is a nitrogen fixing bacterial symbiont of several dozen legume species that has been identified from varying agro-ecological zones (Keyser et al., 1982; Krishnan and Pueppke, 1994). Sinorhizobium sp. were found in three of the five land uses namely; maize-based, shrubland and indeginous forests. These are all land uses likely to have varied species of leguminous plants over time and thus the presence of Sinorhizobium sp. in them was not surprising.
Burkholderia sp. were found in the maize-based system are mostly associated with maize, coffee and sorghum plants (Estrada-De Los Santos et al., 2001). The first isolates of B. vietnamiensiswere recovered from the rhizosphere of rice plants grown in aphytotron (Gillis et al., 1995). As LNB were trapped with siratro, this study shows the wide geographic distribution and substantial capability ofN2-fixing Burkholderia spp. for colonizing diverse host plants.
The differences in the distribution of LNB diversity among the land use types could be attributed to many factors, most important of these being distribution of legumes. Rhizobia are generally host specific (Martinez-Romero and Caballero-Mellado, 1996) and therefore distribution of their hosts is likely to affect their distribution. Planted forests and agroforestry systems, which are land use systems with little or no leguminous plants, were found to have little LNB diversity in comparison with other land use systems that frequently had legumes. Soil physical-chemical properties are also known to affect LNB diversity. For example, Rhizobium populations have been shown to decrease with decrease in soil pH and increase with an increase in pH (Harrison et al., 1989). Soil nitrogen content has also been shown to influence diversity of rhizobia in soils. High levels of nitrogen in the soil have been shown to decrease the diversity of rhizobia in the soil (Hirsch, 1996; Palmer & Young, 2000).
Whereas effort was not focused on measuring these factors, it was clear that the different landuse systems were subjected to different soil amendment practices. Soil amendments such as addition of manure, lime and fertilizers do affect soil pH and soil nitrogen content, thereby influencing diversity of LNB. Lastly, Venkateswarlu et al. (1997) suggested that the abundance of native rhizobial population is also heavily influenced by 'crop related factors'. These crop related factors such as production of root exudates by plants and quality of organic matter returned to the soil by plants could also have contributed to the differences in LNB diversity among the land uses.
However the isolates obtained from this study may not reflect the true diversity of LNB in Taita-Taveta because of two key reasons: (i) only one host trap (siratro) was used, and (ii) a soil dilution series were used to trap the indigenous LNB. Siratro is often the host of choice for trapping slow-growing, alkali-producing LNB (Somasegaran and Hoben, 1994), but results from this study demonstrated that, contrary to this general opinion, the trap plant attracted fast-growing, acid- producing types as well. Bala et al. (2001) showed that soil dilutions may select for certain types/genotypes depending on the dilution level and differences between strains in their abundance, competitiveness for nodule formation and the potential influence of the soil environment on competitive success. It may therefore be prudent to, in the future, use trap hosts with varying LNB affinities, and also explore nodulation in situ where possible, to evaluate the diversity of indigenous or naturalized symbiotic populations.
CONCLUSION
The diversity of LNB varied with the different fanning systems in Taita-Taveta. LNB populations in the soils were variable, in terms of both abundance and the symbiotic efficiencies. Results also demonstrated a high population of rapid-growing rhizobia in the soils studied. LNB were classified into six genera namely; Agrobacterium, Rhizobium, Bradyrhizobium, Sinorhizobium, Burkholderia and Herbaspirillum, based on RFLP and sequencing of 16 S rRNA genes.
ACKNOWLEDGEMENTS
The authors wish to acknowledge financial support from Conservation and Sustainable Management of Belowground Biodiversity (CSM-BGBD) Project through funds from GEF/UNEP. Appreciation also goes out to Kenya Forestry Research Institute and Jomo Kenyatta University of Agriculture and Technology for providing laboratory facilities.
REFERENCES
Anyango, B., Keya, S., and Owino, F. 2005. Occurence of Nodulation in Leguminious Trees in Kenya. Journal of Tropical Microbiology and Biotechnology. 1: 21-26. [ Links ]
Anyango, B., Wilson, K. I, Beynon, J. L. and Giller, K. E. 1995. Diversity of rhizobia nodulating Phaseouls vulgaris L. in two Kenyan soils with contrasting pHs. Applied and Environmental Microbioliogy. 61: 4016-4021. [ Links ]
Bala, A., Murphy, P. and Giller, K.E., 2001. Genetic diversity of rhizobia from natural populations varies with the soil dilution sampled. Soil Biology and Biochemistry. 33: 841-843. [ Links ]
Bala, A., Murphy, P.J., and Giller, K. E. 2004. Classification of tropical tree rhizobia based on phenotypic characters forms nested clusters of phylogenetic groups. West African Journal of Applied Ecology. 6: 9-19. [ Links ]
Baldani, J. I., Baldani, V. L. D., Seldin, L. and Dobereiner, J. 1986. Characterization of Herbaspirillum seropedicae gen. nov., sp. nov., a root-associated nitrogen-fixing bacterium. International Journal of Systematic Bacteriology. 36: 86-93. [ Links ]
Bennett, J. E., Woomer, P. L. and Yost, R. S. 1990. Users manual for MPNES most-probable-number enumeration system ver. 1.0. NifTAL project and University of Hawaii. [ Links ]
Broughton, W. J. And Dilworth, M. J. 1971. Control of leghaemoglobin syntheisis in snake beans. Biochemical Journal. 125: 1075-1080. [ Links ]
Bumpus, E. D. 1957. Legume nodulation in Kenya. East African Agricultural and Forestry Journal. 23:91-99. [ Links ]
Cheng, F., Cao, G., Wang, X., Zhao, J., Yan, X. And Liao, H. 2009. Isolation and application of effective nitrogen fixation rhizobial strains on low-phosphorous acid soils in South China. Chinese Science Bulletin. 412-420 [ Links ]
Estrada-De Los Santos, P., Bustillos-Cristales, R. and Caballero-Mellado, J. 2001. Burkholderia, a Genus Rich in Plant-Associated Nitrogen Fixers with Wide Environmental and Geographic Distribution. Applied and Environmental Microbiology. 67: 2790-2798. [ Links ]
Felsenstein, J. 1985. Confidence limits on phytogenies: An approach using the bootstrap. Evolution. 39: 783-791. [ Links ]
Gibson, A. H. 1987. Evaluation of nitrogen fixation by legumes in the greenhouse and growth chamber. In A. H. Gibson, Symbiotic Nitrogen Fixation Technology (pp. 321-363). New York: Marcel Dekker. [ Links ]
Gillis, M., Tran Van, V., Bardin, R., Goor, M., Hebbar, P., Willems, A., Segers, P., Kersters, K., Heulin, T. and Fernandez, M. P. 1995. Polyphasic taxonomy in the genus Burkholderia leading to an emended description of the genus and proposition of Burkholderia vietnamiensis sp. nov. for N2-fixing isolates from rice in Vietnam. International Journal of Systematic Bacteriology. 45:274-289. [ Links ]
Harrison, S., Jones, D., Young, J. P. 1989. Rhizobium population genetics: genetic variation within and between populations from diverse locations. Journal of General Microbiology. 135: 1061-1069. [ Links ]
Hirsch, P. R. 1996. Population dynamics of indigenous and genetically modified rhizobia in the field. New Phytologist. 133: 159-171. [ Links ]
Hurek, T., Wagner, B. and Reihold-Hurek, B. 1997. Identification of N2-fixing plant-and fungus-associated Azoarcus species by PCR-based genomic fingerprints. Applied and Environmental Microbiology. 63: 4331-4339. [ Links ]
Karanja, N K. Ayuke, F.O., Muya, E. M., Musombi, B. K. and Nyamasyo, G. H. N 2009. Soil macrofauna community structure across land use systems of taita, kenya. Tropical and Subtropical Agroecosystems. 11: 385 - 396. [ Links ]
Keyser, H. H., Bohlool, B. B., Hu, T. and Weber, D. F. 1982. Fastgrowing rhizobia isolated from root nodules of soybean. Science. 215: 1631-1632. [ Links ]
Khbaya, B., Neyra, M., Normand, P., Zerhari, K., and Filati-Maltouf, A. 1998. Genetic diversity and phlogeny of rhizobia that nodulate Acacia spp. in Morrocco assessed by analysis of rRNA genes. Applied and Environmental Microbiology. 64: 4912-4917. [ Links ]
Krishnan, H. B. and Pueppke, G. 1994. Host range, RFLP, and antigenic relationships between Rhizobium fredii strains and Rhizobium sp. NGR234. Plant Soil. 161: 21-29. [ Links ]
Laranjo, M, Rodrigues, R., Alho, L., and Oliviera, S. 2001. Rhizobia of Chickpea from Southern Portugal: symbiotic efficiency and genetic diversity. Journal of Applied Microbiology. 90: 662-667. [ Links ]
Leonard, L. T. 1943. A simple assembly for use in testing of culture of rhizobia. Journal of Bacteriology. 45: 523-527 [ Links ]
Martinez-Romero, E., and Caballero-Mellado, J. 1996. Rhizobium phytogenies and bacterial genetic diversity. Critical Reviews in Plant Sciences. 15: 113-140. [ Links ]
McDonald, J. 1935. The inoculation of leguminous crops. East African Agricultural and Forestry Journal. 1: 8-13. [ Links ]
Morrison, J. 1966. Productivity of grass and grass/legume swards in Kenya highlands. East African Agricultural and Forestry Journal. 32: 19-24. [ Links ]
Ngokota, L., Krasova-wade, T., Etoa, F. X., Sylla, D., and Nwaga, D. 2008. Genetic diversity of rhizobia nodulating Arachis hypogaea L. in diverse land use systems of humid forest zone in Cameroon. Applied Soil Ecology. 40: 411-416. [ Links ]
Odee, D. W., Haukka, K., Mclnroy, S. G., Sprent, J. I., Sutherland, J. M. and Young, J. P. W. 2002. Genetic and symbiotic characterization of rhizobia isolated from tree and herbaceous legumes grown in soils from ecologically diverse sites in Kenya. Soil Biology and Biochemistry. 34: 801-811. [ Links ]
Odee, D. W., Sutherland, J. M., Kimiti, J. M. And Sprent, J. I. 1995. Natural populations and nodulation status of woody legumes growing in diverse Kenyan conditions. Plant and Soil. 173:211-224. [ Links ]
Odee, D. W., Sutherland, J. M., Makatiani, E. T., Mclnroy, S. G. and Sprent, J. I. 1997. Phenotypic characteristics and composition of rhizobia associated with woody legumes growing in diverse Kenyan conditions. Plant and Soil. 188: 65-75. [ Links ]
Olivares, F. L., Baldani, V. L. D., Reis, V. M., Baldani, J. I. and Döbereiner, J. 1996. Occurrence of the endophytic diazotrophs Herbaspirillum spp. in roots, stems, and leaves, predominantly of Gramineae. Biology and Fertility of Soils. 21: 197-200 [ Links ]
Palmer, K. M. and Young, J. P. W. 2000. Higher diversity of Rhizobium leguminosarum Biovar viciae in arable soils than in grass soils. Applied and Environmental Microbiology. 66: 2445-2450. [ Links ]
Paul, E. A. and Clark, F. E. 1989. Soil microbiology and biochemistry. E.A Paul, F.E Clark Academic Press, San Diego [ Links ]
Saitou, N., and Nei, M. 1987. The neighbour-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 4: 406-425. [ Links ]
Sambrook, J., Fritsch, E. F. and Maniatis, T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [ Links ]
Sawada, H., Kuykendall, D., and Young, J. 2003. Changing concepts in the systematics of bacterial nitrogen-fixing legume symbionts. Journal of General and Applied Microbiology. 49:155-179. [ Links ]
Shayne, J. J., Hugenholtz, P., Sangwan, P., Osborne, C. and Jansen, H. P. 2003. Laboratory cultivation of widespread and previously uncultured bacteria. Applied and Environmental Microbiology. 69: 7211-7214. [ Links ]
Somasegaran, P. and Hoben, H. J. 1994. Handbook for Rhizobia: Methods in Legame-Rhizobium technology. Berlin: Springer-Verlag. [ Links ]
Souza, D. I. A. 1969. Legume nodulation and nitrogen fixation studies in Kenya. East African Agricultural and Forestry Journal. 34: 299-305. [ Links ]
Tamura, K., Dudley, J., Nei, M. and Kumar, S. 2007. MEGA4: Molecular Evolutionary genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 24: 1596-1599. [ Links ]
Tamura, K., Nei, M. and Kumar, S. 2004. Prospects for inferring very large phytogenies by using the neighbour-joining method. National Academy of sciences Conference (pp. 11030-11035). New York: National Academy of Sciences. [ Links ]
Valverde, A., Velazquez, E., Gutiérrez, C, Cervantes, E., Ventosa A. and Igual, J. 2003. Herbaspirillum lusitanum sp. nov., a novel nitrogen-fixing bacterium associated with root nodules of Phaseolus vulgaris. International Journal of Systematic and Evolutionary Microbiology. 53:233-239. [ Links ]
Venkateswarlu, B., Hari, K. And Katyal, J. C. 1997. Influence of soil and crop factors on the native rhizobial populations in soils under dryland fanning. Applied Soil Ecology. 7: 1-10. [ Links ]
Vincent, J. M. 1970. A manual for the practical study of root-nodule bacteria. Blackwell, Oxford. [ Links ]
Zawedde, J., Rwakaikara, M. C, Kizza, C, Lamtoo, G., and Okwakol, M. 2009, June 19. Rhizobia. Retrieved November 12, 2009, from www.bgbd.or.ug: http://www.bgbd.or.ug [ Links ]
Zewdu, T., Giselle, N., Sini, S., Lars P., and Kristina L. 1998. Phytogeny ofRhizobium galegae with respect to other rhizobia and agrobacteria. International Journal of Systematic Bacteriology. 48: 349-356. [ Links ]
Zhang, X., Harper, R., Karsisto, M, and Lindstrom, K., 1991. Diversity of Rhizobium bacteria isolated from the root nodules of leguminous trees. International Journal of Systematic Bacteriology. 41: 104-113. [ Links ]














