Introduction
In the last two decades, worldwide interest has increased in the study of various biological objects, such as cells, tissues, and microorganisms, using the method of secondary ion mass spectrometry (SIMS) [1-15]. Sputtering of the analyzed surface with simultaneous registration of secondary ions makes it possible to obtain the distribution of any elements of the Periodic Table and any stable isotope on the analyzed surface with record sensitivity. And continuous ion sputtering allows obtaining a three-dimensional distribution of elements of interest. The new generation of time-of-flight mass spectrometers are equipped with pulsing liquid-metal sources of heavy ions of bismuth or gold, allowing to achieve a lateral resolution of about 100 nm. In addition, irradiation with heavy ions of organic materials leads to a rather high secondary ion emission, that is, to a high sensitivity of the analysis. On the other hand, the widespread use of the SIMS method in biology and medicine is still limited by the lack of an effective sample preparation technique. Mass spectrometers operate with a high vacuum in the analysis chamber (10-10 - 10-9 Torr), which imposes special requirements on the sample under study: first, the sample must be solid and must not emit gas or water molecules. Therefore, dehydration of biological samples is one of the prerequisites [2-4,8]. Cryo-sectioning of frozen samples is one of the possible alternatives for sample preparation and analysis by SIMS method [4,11-15]. At the same time, despite lengthy research, there is still no single protocol for preparing bio samples for the SIMS method. A preparation method successfully applied to one sample, often shows poor results in others.
In this article, we demonstrate the experimental results of an elemental analysis performed by SIMS for a set of mouse fibroblasts cells from the established 3T3F442A line, which could differentiate into adipocytes under certain culture conditions. The cells were cultured, fixed, and dehydrated using two different reagents and sequential purification methods. At the same time, all the specific problems of SIMS analysis of cell culture and other bio-samples are discussed in the work.
Experimental
Two groups of 3T3-F44A2 fibroblasts were seeded at 1.25×103 cells/cm2 on crystalline silicon substrates in adipogenic medium consisting of DMEM supplemented with 5% bovine calf serum, 5 μg/mL insulin, and 1 μM d-biotin until the fatty phenotype was achieved [16,17]. Eight days later, cells were fixed: one group with 2% glutaraldehyde in PBS (137 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, and 2 mM KH2PO4), the other group in 3.5% paraformaldehyde in PBS for 10 minutes. Both groups were preserved in 0.9% NaCl. After fixation, the samples were washed in deionized water several times (up to three times), wash results were compared. Then the samples were dried in a nitrogen atmosphere. Two series of experimental samples were prepared using both chemicals: we used the first series to study the effect of washing and the second to study the distribution of elements within cells to compare the effect of the chemicals used.
SIMS analysis of the lateral distribution of the selected elements of cell culture was performed by using a TOF-SIMS-V instrument from Ion-TOF GmbH. A pulsed finely focused bismuth ion beam with an energy of 30 keV and a beam diameter of about 150 nm was used. First, the analysis area was selected using a secondary electronic image obtained by scanning a surface area with a size of 500×500 μm2 under irradiation with Bi+ ions. After that the surface was sputtered with a Bi+ ion beam in the so-called direct current mode to remove surface contaminations and cell membrane. Then, during the analysis, secondary ions were recorded sputtered from the analyzed surface with sizes from 50×50 µm2 to 100×100 µm2. Secondary positive ions were monitored to analyze the distribution of metallic and semimetallic elements in cells, and negative secondary ions were used to analyze the distribution of halogens and non-metals. So, two different analyzes of the same area were carried out. Only one stable isotope of the element of interest with the maximum concentration was analyzed each time. For the analysis of elements such as nitrogen (with a low secondary ion yield of both positive and negative ions), secondary cluster ions like CN- were used. Secondary ion signals were accumulated during 50-100 cycles to get better statistics and obtain quality ion images.
Experimental results and discussion
The culture medium contains a variety of amino acids, minerals, and salts that enable cell growth and differentiation; residues of these components can reside on the cell surface. In addition, the fixatives used are dissolved in PBS, which contains salts necessary to maintain cell osmolarity. Therefor the sample preparation process included the procedure of their washing with deionized distilled water after fixation. Figure 1 shows ion images of cells fixed with p-formaldehyde on the surface of a silicon crystal after single (a) and double (b) washing in DI water. The area of the ion image is 50×50 µm2, images are shown in secondary positive ions (from left to right and top to bottom): Na+, Si+, K+, CaO2 +. As can be seen from the distributions obtained, a single wash does not remove all external elements on the surface of cells, such as sodium present in the fixative reagents. Moreover, the surface contaminant (see the ion image with Na+, K+, and CaO2 + ions) prevents the observation of the cell structure. So, multiple washes of the prepared sample are necessary to obtain a clean surface with a preserved cell structure. Subsequently, the samples were washed three times.
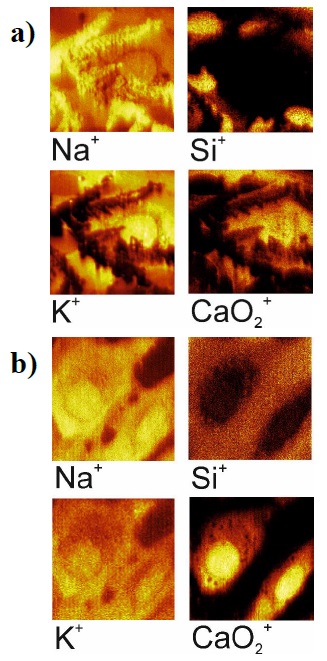
Figure 1 Ion images (50×50 µm2) of the cells fixed with paraformaldehyde after a single (a) and double (b) washing.
There are a number of reagents that can be used for tissue fixation. The most popular are formaldehyde, used for histopathology and other technical studies, and glutaraldehyde, used for ultra-structural studies. Formaldehyde reacts with protein side chains to form reactive hydroxy-methyl groups. It can penetrate nuclear proteins and nucleic acids, stabilizing the nucleic acid-protein envelope and modifying nucleotides by reacting with free amino groups. Glutaraldehyde or glutaric dialdehyde (CHO(CH2)3CHO) is considered a bifunctional aldehyde that has aldehyde groups at both ends of the molecule that can react with the same chemical groups as formaldehyde. Tissues or cells fixed in glutaraldehyde will cross-link more intensely than tissues fixed in formaldehyde. We use both chemicals to fix cells after their dehydration.
Figure 2a shows images of cells obtained in an optical microscope (right image, ×10 magnification) and in secondary electrons emitted during ion irradiation (left) (a), as well as secondary negative ion images with (b): H-, CH-, O-, F-, CN-, Si-, PO2 -, Cl-, and total ions for cells prepared with glutaraldehyde. The optical images include green square which correspond to 500×500 µm2 area (as maximal analyzed area); the red square shows the analyzed area. These figures show well-defined cells with cytoplasm stretched in silicon, a well-defined nucleus, and no cellular damage. Near the nucleus and in the cytoplasm, electrodense areas with H-, CH-, CN- and Cl- are observed.
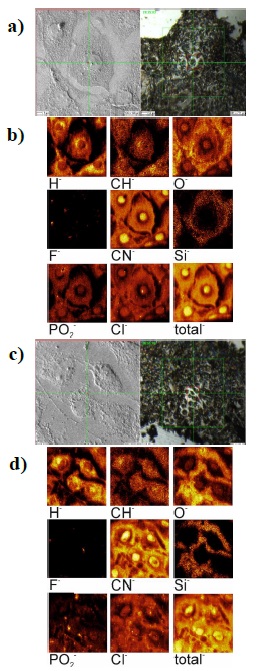
Figure 2 Image of cells fixed after dehydration with glutaraldehyde (a, b) and with paraformaldehyde (c, d); a) and c) show electronic and optical images; b) and d) show ionic images (100×100 µm2).
Figure 2 (c, d) shows for comparison cells prepared using paraformaldehyde. Optical and secondary electronic images (c) and images of the distribution of various ions (d) are also shown, the same as for the previous series of image. These images show cell damage, retracted cytoplasm, reduced cell volume, subcellular localization of ions is not clear.
Based on the images obtained, several conclusions can be drawn:
- The use of paraformaldehyde leads to cell damage after dehydration. When glutaraldehyde is used, the cell structure is completely preserved (the shape and volume of the cells), as seen in Figure 2 (a, b). This may be due to the fact that glutaraldehyde replaces more of the substance in the cells after dehydration, thereby preserving the cell structure.
- The resulting lateral resolution (on the order of 200 nm) allows us to visualize the basic structure and shape of the cells under study. The maximum lateral resolution of TOF-SIMS in the ion images mode is approximately 100 nm. As can be seen from the images in Cl- and PO2 - ions, it is quite possible to study the internal structure of cells for the distribution of chemical elements in cellular elements (nucleus, vacuoles, Golgi, etc.). But this would require 3-D analysis, with sequential removal of cell material and visualization of underlying layers. Such analysis will be realized in our future study.
- Unfortunately, some elements are introduced by the used reagents and water in sufficiently large quantities and washing the samples does not allow them to be removed. First, we are talking about sodium, potassium (not shown in the figures), chlorine and calcium, which are interesting in terms of studying cellular metabolism. In the case of most other biometals: Mg, Al, Fe, Cu, and Zn, there should be no such problem, that is, the distribution of these elements can be studied. This is important because transition metals are key components of numerous proteins and enzymes involved in electron transport [18]. For example, cuprate plays an important role in the function of many enzymes that produce neurotransmitters in the central nervous system, as well as other enzymes critical for electron transport and energy production in mitochondria [19]. In addition, alimentary or genetically determined deficiencies of these metals are associated with various diseases, such as anemia, hypoxemia, and other pathological disorders.
Of course, if stable isotopes are used during cell growth, an effective analysis of their distribution in cells after biochemical reactions is also possible.
- As for all other methods of sample preparation, quantitative analysis of elements according to the obtained ion distributions is not yet possible. That is, quantitative estimates are possible only for experiments with stable isotopes (in comparison with the normal distribution of isotopes).
We also conducted a study of the “stability” of the prepared cell samples over a fairly long period. Figure 3 shows images of an individual cell prepared (fixed) using glutaraldehyde two and a half years after the preparation. All this time, the sample was stored in the refrigerator at a temperature of -4 °C. As for the previous experiments, optical and secondary electronic images (a) and ion images (b) are given. Secondary negative ions (from left to right and from top to bottom) were recorded: H-, O-, CN-, Si-, Cl- and NaO-. The structure of the cell is partially destroyed, but some elements, such as the cell nucleus, can still be studied for chemical composition. Based on the obtained images, we can conclude about quit a high stability of the cells prepared by this method.
Conclusions
Standard methods of biological cell preparation, including glutaraldehyde and paraformaldehyde fixation, make it possible to study cells directly grown on the surface of crystalline silicon by three-dimensional SIMS analysis of element distribution in these cells. Due to greater penetration into the cell, glutaraldehyde allows the maximum preservation of the volume and shape of the cell, including cell components (at least the cell nucleus). The cells prepared by this method shows a very good stability with time, necessary for analysis by SIMS and by other physical methods.
Analysis of the distribution of elements, including stable isotopes used for example in drugs, in elements of cell structures: nucleoli, vacuoles, Golgi and even in the membrane, requires a three-dimensional SIMS analysis. A methodology for such an analysis is being developed within our research group and will be published later.
The problem of quantitative analysis of elements, which is inherent to the SIMS method, can be partially solved by using stable isotopes in the process of growing cells of interest to study the metabolism of individual cells or to study the effects of medications at the cellular level.











 nova página do texto(beta)
nova página do texto(beta)




