INTRODUCTION
HFF-related Hemochromatosis (HH) is the most common inherited iron overload disease in Caucasian populations. Homozygosity for the p.C282Y mutation in HFE is the typical defect conferring a strong predisposition to develop the disease.1 It leads to inadequate production of hepcidin, the master regulator of iron homeostasis through a still incompletely understood mechanism.1 Several evidences indicate that p.C282Y homozygosity has variable penetrance and expression. While most homozygotes for the p.C282Y mutation show alteration of serum
iron indices, severe iron overload and iron-related complications are observed in less than 30% of men and 5% of women.2 - 4 Alcohol intake and obesity, and coexistence with other diseases able to increase iron absorption or favour liver damage, can influence phenotype expression.5 - 9 ) The role of genetic modifiers was demonstrated in HFE- knock-out mice,10 but association studies between genetic markers and disease phenotype in humans have given conflicting results.11 , 16 Recently, McLaren, et al. 17 identified glyceronephosphate O-acyltransferase (GNPAT) rs11558492 A>G polymorphism (p.D519G) as a new potential genetic modifier of HFE hemochromatosis phenotype in p.C282Y homozygotes. The prognostic role of this polymorphism has been evaluated in other cohorts,18 - 20 but none demonstrated an association between the SNP and markers of iron overload. However, Besson-Fournier, et al. found allele G to be significantly enriched in the subset of males with markedly increased iron stores. More recently, in a small cohort of healthy women and in 59 individuals not carrying HFE genotypes 8 hours after iron challenge an association between minor (G) GNPAT allele and serum iron was reported.21 , 22 To contribute in understanding the role of this SNP as a possible modifier of iron phenotype, we analysed alleles and genotypes frequencies and their relationship with iron parameters in a cohort of298 HFE p.C282Y homozygotes of Italian ancestry and a group of 169 male healthy blood donors.
MATERIAL AND METHODS
Patients
Two hundred and ninety-eight patients (205 men and 93 women) homozygous for p.C282Y in HFE with a median age of 48 years (interquartile range (IQR) 36-57 years) were studied. They attended the Centre for Disorder of Iron Metabolism at ASST-Monza-S.Gerardo Hospital, Fondazione IRCCS Ca’ Granda Policlinico Hospital in Milan and Policlinico GB Rossi in Verona. Patients were selected based on availability of serum ferritin (SF), transferrin saturation (TS), alcohol intake and serum alanine aminotransferase (ALT) at diagnosis, and good quality DNA. Patients with coexistent chronic liver diseases (alpha-1 antitrypsin deficiency, autoimmune hepatitis, chronic B or C virus hepatitis), and alcohol intake > 50 g/day in men and >30 g/day in women23 were excluded.
Two-hundred and twenty-six patients (160 men and 66 women) underwent to liver biopsy for diagnostic (before HFE testing) and/or prognostic aim. Liver fibrosis was assessed according to Ishak, et al.24 Liver iron concentration (LIC) was available in 191 (64%) patients and was measured on liver biopsy samples in fresh or deparaffinized specimens (N = 155) or by quantitative magnetic resonance (MR) as previously reported.25 , 26 The lower number of available LIC is explained by the fact that:
Iron removed (IR) after iron depletion was available in 234 (78.5%) patients and measured as previously reported23
One hundred and sixty-nine Italian health blood donors, selected from a cohort previously studied to define prevalence of HFE mutations in Italy,27 were also evaluated as controls. Selection was based on HFE genotype (only wild-type or p.H63D heterozygotes were retained), sex (females were excluded), availability of haemoglobin and SF at first blood donation, and good quality DNA.
Patients gave their informed consent for liver biopsy and both patients and controls for using their DNAs previously collected for HFE testing according to Institutions’ policy. The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki as reflected in a priori approval by the appropriate institutional review committee.
GNPAT polymorphism analysis
Genomic DNA of patients and controls was extracted from peripheral blood leucocytes using the Wizard® Genomic DNA Purification kit (Promega, Madison, WI, USA) or the ZR DNA-Card Extraction Kit™ (Zymo Research, Irvine, CA, USA) according to manufacturer’s instructions. GNPAT rs 11558492 genotyping was carried out by tetra-primer Amplification Refractory Mutation System (ARMS-PCR) and verified by 1.5% of agarose gel stained with ethidium bromide, and/or TaqMan® 5’-nuclease assays (LifeTechnologies, Carlsbad, CA). Random samples were confirmed by direct sequencing.
Statistical analysis
Median and IQR were used for descriptive purposes as appropriate. Hardy-Weinberg equilibrium was examined for all subjects using χ2 test. To evaluate the prognostic role of GNPAT polymorphism, we evaluated allele and genotype frequencies according to TS, SF, LIC and IR levels in patients and SF levels in controls. In patients, analyses were performed in the whole series and in males only. A further analysis was done comparing subgroups of male patients with extreme phenotypes: severe iron overload (SF > 2,000 μg/L, IR > 10 g and/or LIC > 250 μmol/ g, alcohol intake < 30 g/day), mild iron overload (SF < 1000 μg/L, IR < 5 g and/or LIC <100 μmol/g, alcohol intake < 30 g/day). To evaluate the effect of GNPAT rs11558492 on liver fibrosis we grouped patients in two groups: those with absent or mild-moderate fibrosis (stage 0-3) and with severe fibrosis-cirrhosis (stage 4-6) as previously reported.23 Comparisons of GNPAT rs11558492 allele and genotype frequencies between groups were done by Fisher’s exact test. Statistical analyses were performed using GraphPad Prism software, version 4.0 (San Diego, CA, USA). P-value < 0.05 was the accepted level of significance.
RESULTS
Genotype distributions of GNPAT rs11558492 were in Hardy-Weinberg equilibrium in patients and controls. Table 1 shows main data of HFE-HH patients and controls. Serum ferritin significantly correlated with LIC (R2 = 0.42; p < 0.0001) and IR (R2 = 0.6; p < 0.0001), and LIC with IR (R2 = 0.51; p < 0.0001). As shown in table 2, genotype and allele frequencies were comparable in patients and healthy blood donors, and those reported in the Exorne Variant Server (EVS) database (genotypes: AA=64%; AG = 31.2%; GG = 4.8%; alleles: A = 79.7% and G = 20.3%).
Table 1 Main data of HFE-HH patients and controls. Data are expressed as median (interquartile range).
† Iron removed available in 234 out of 298 (78.5%). †† Liver iron concentration was available in 191 out of 298 (64 %). NA: not available.
Table 2 Genotype and allele frequencies (number and percentage) of GNPAT rs11558492 polymorphism in HFE-HH patients (whole cohort and male-only) and healthy blood donors. Data are expressed as numbers and (percentages)
No significant difference between patients and controls.
Figure 1 A-C shows median and IQR of SF according to GNPAT genotypes in the whole cohort [AA: 1,000 μg/L (552-1815.5); AG + GG: 1061 μg/L (610.5-1946.5)] and male-only [AA: 1145 μg/L (801-2019); AG + GG: 1245 μg/L (771-2721)] HH patients, and in controls [AA: 78.0 μg/L (55.5-117.5); AG + GG: 83.0 μg/L (53-110)]. No significant difference was observed. Accordingly, also LIC and IR did not differ according to GNPAT genotypes both in the whole cohort of HH patients [LIC: AA: 214.2 μmol/g (158.7-361.2); AG + GG: 242.6 μmol/g (167.2-310.2); IR: AA: 6.25 g (4-9.9); AG + GG: 5.77 g (3.9-10)] and in male-only subgroup [LIC: AA: 220 μmol/g (167.2-365.1); AG + GG: 263.1 μmol/g (179.4-320); IR: AA: 8 g (5-12); AG + GG: 8 g (4.7- 11.7)] (Figures 2 A-B and 3 A-B). No differences were also observed for TS [AA genotype: 85% (72-91.6); AG + GG genotypes: 90% (76.7-97.5)], haemoglobin and ALT (data not shown). We then measured the frequency of GNPAT alleles and genotypes in the subgroups of patients with extreme phenotypes. Thirty and thirty- eight men fulfilled the criteria for severe and mild iron overload, respectively. Allele and genotype frequencies did not differ between the two subgroups (Table 3) and compared to EVS database. Fifty-eight out of 226 patients had severe fibrosis/cirrhosis. Minor allele frequency (MAF) was slightly higher in patients with severe fibrosis/cirrhosis (25%) than in those with absent/mild fibrosis (20.5%), but not significantly so (p = 0.36). Similarly, in the subgroup of men with alcohol intake < 30 g/day, MAF was 25% vs. 19.1%, respectively (p = 0.31).
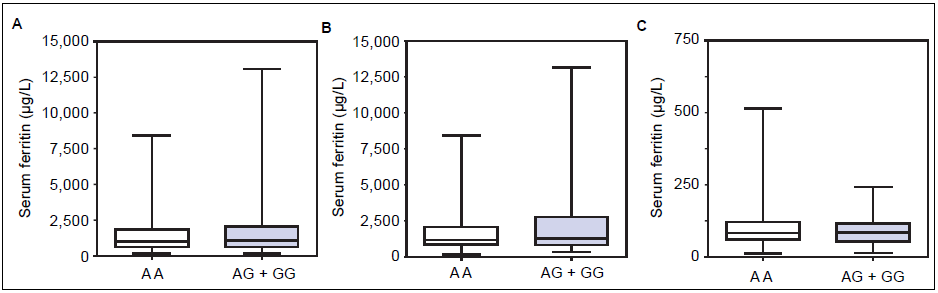
Figure 1 Serum ferritin levels according to GNPAT genotypes in hemochromatosis (HFE-HH) patients (whole cohort) (A), hemochromatosis male-only patients (B) and healthy blood donors (C).
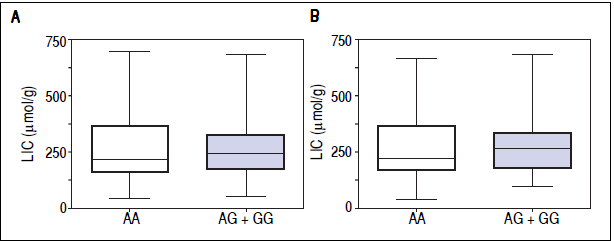
Figure 2 Liver iron concentration according to GNPAT genotypes in hemochromatosis (HFEHH) patients (whole cohort) (A) and hemochromatosis male-only patients (B).

Figure 3 Iron removed according to GNPAT genotypes in hemochromatosis (HFE-HH) patients (whole cohort) (A) and hemochromatosis male-only patients (B)
Table 3 Allele and genotype frequencies in men with HH selected according to iron phenotype. Data are expressed as numbers and (percentages).
Severe iron overload: SF > 2000 μg/L, IR >10 g and/or LIC > 250 μmol/g, alcohol intake < 30 g/day. Mild iron overload: SF < 1,000 μg/L, IR < 5 g and/or LIC < 100 μmol/g, alcohol intake < 30 g/day).
DISCUSSION
We evaluated the frequency of GNPAT alleles and genotypes in a cohort of Italian p.C282Y homozygous patients and healthy blood donors and the relation between the SNP and markers of iron status. The frequency of alleles and genotypes in patients did not differ compared to healthy blood donors and those reported in EVS database. Allele frequencies of GNPAT p.D519G were slightly lower than that reported in other series of HH patients originating from Western Europe,18 - 20 but similar to that of European Americans from the EVS database. Also, we did not find associations between alleles and genotypes and TS, SF, LIC and IR in HFE-HH patients, and SF in blood donors. Our results contrast those reported by McLaren, et al.17 in North-American and Australian HH patients, but agree to those reported in other cohorts18 , 19 , 28 of HFE-HH patients. Also, we could not find differences comparing allele and genotype frequencies in the subsets of HFE-HH patients selected for mild and severe iron phenotypes. We used restrictive criteria (SF > 2,000 μg/L and IR > 10 g and/or LIC > 250 μmol/g) to define severe iron phenotype in HFE-HH, but similar results were observed even using a ferritin cut-off of 1,000 μg/L instead of 2,000 μg/L (N = 67 men with alcohol intake < 30 g/day; MAF: 23.1%).
McLaren, et al.7 linked controversies to the design and power of studies and to inclusion criteria and supported the role of GNPAT as a modifier of iron phenotype showing that it might participate in hepcidin regulation through a still undefined mechanism in vitro. We hypothesised that analysing subjects of Italian ancestry whose genetic structure is heterogeneous might help in shedding light on these discrepancies.29 Our findings indicate that GNPAT rs11558492 is not a major modifier of iron status in HFE- HH, although we cannot exclude that it could be part of a more complex genetic background that might include more than one mild modifier.11 , 16 Accordingly, we were not able to show any relation between GNPAT polymorphism and iron status in healthy blood donors. The latter finding contrast those by Hsiao, et al. 21 who showed an association between serum iron at baseline and after an iron test in 83 young women. Similarly, Rametta, et al 22 demonstrated that carriers of the G variant without HFE genotypes at risk (p.C282Y wild type or p.H63D heterozygotes) have mildly increased iron absorption 8 hours after ferrous sulphate challenge. However, both studies did not find association between GNPAT and SF that represents the best marker of iron stores in healthy people. These results raised further questions on the clinical relevance of GNPAT as iron modifier. Last, GNPAT rs1155849 was not associated with severe fibrosis/cirrhosis, confirming previous results.18 In conclusion, several evidence indicate that homozygosity for p.C282Y in HFE is necessary but not sufficient to develop significant iron overload. There is general agreement that acquired factors, such as alcohol intake, hepatic steatosis, obesity and coexistence of chronic viral hepatitis, are major modifiers of iron or clinical phenotype in HH.5 - 9 Conversely, though milder effects cannot be ruled out in the absence of adequately powered cohorts, no polymorphisms that came up from studies done up to now seem to exert a major effect on iron phenotype to be considered for defining genetic susceptibility to fully penetrant HFE-HH in clinical practice. Interestingly, two recent studies strongly suggested a role for rs236918 in PCSK7 gene as a modifier of risk of liver fibrosis in p.C282Y homozygotes.23 , 30 Exorne or whole-genome sequencing studies by next-generation sequencing technologies will enable to further explore if there exist some other unknown genetic variants with clinical significance in HFE-HH patients carefully selected and controlled for environmental factors.
ABBREVIATIONS
ALT: |
Serum alanine aminotransferase. |
ARMS-PCR: |
Amplification refractory mutation system - polymerase chain reaction. |
EVS: |
Exorne variant server. |
GNPAT: |
Glyceronephosphate O-acyltransferase. |
HH: |
Hereditary hemochromatosis |
IQR: |
Interquartile range. |
IR: |
Iron removed. |
SF: |
Serum ferritin. |
SNP: |
Single nucleotide polymorphism. |
TS: |
Transferrin saturation. |











 nueva página del texto (beta)
nueva página del texto (beta)


