Introduction
Maize (Zea mays L.) is one of the most important staple foods in developing countries, but it has a poor protein quality due to the deficiency of lysine (Lys) and tryptophan (Trp). The maize opaque2 mutation typically doubles the Lys and Trp contents of wild-type kernels (Mertz et al., 1964). However, the agronomic problems associated with the soft endosperm of this mutant limited its use. Plant breeders developed hard-kernel o2 genotypes known as Quality Protein Maize (QPM) (Villegas et al., 1992), but the mechanism by which the soft endosperm of o2 becomes hard and vitreous in QPM is not entirely understood. Some studies have demonstrated an essential role for the 27-kDa γ-zein in endosperm modification (Wu et al., 2010; Liu et al., 2016); the enhanced accumulation of 27-kDa γ-zein favors the formation of protein bodies and their compaction between starch granules. Gibbon et al. (2003) proposed a complementary mechanism where alterations in starch structure facilitate that granules associate with one another, and the spaces between them are filled in the vitreous areas of the kernel.
Genetic analysis with recombinant inbred lines (RILs) derived from a cross between W64Ao2 and K0326Y QPM identified three quantitative trait loci (QTL) in bins 1.06, 7.02, and 9.03 associated with endosperm modification (Holding et al., 2011). The locus in bin 7.02 was linked to the 27-kDa γ-zein, whereas that in bin 9.03 was close to starch biosynthesis genes. Salazar-Salas et al. (2014) evaluated the effect of each QTL on protein synthesis and starch physicochemical properties in the mature endosperm. They found that the vitreous phenotype of QPM is associated with the synthesis of less crystalline starch containing more amylose and short-intermediate amylopectin branch chains, which may favor starch granules packing. However, knowledge about the genetic and molecular basis of these changes in starch composition is scarce. This study aimed to identify QTL associated with starch physicochemical properties, and to evaluate the expression of starch biosynthesis genes and its relationship with starch composition in developing endosperm of maize lines contrasting in vitreousness and the genotype of QTL flanking markers.
Materials and methods
Plant materials
Maize inbred lines K0326Y QPM (vitreous) and W64Ao2 (opaque) and 103 recombinant inbred lines (RIL) derived from their cross were used. They were grown at the Faculty of Agronomy field station in Culiacan, Sinaloa, Mexico (24°37’29” N, 107°26’37” W), in 2015. Kernels were degermed and ground into a fine powder. Based on QTLs previously identified for endosperm modification (Holding et al., 2011), six RILs contrasting in vitreousness and the genotype of flanking markers were selected. Three vitreous RILs were homozygous for the K0326Y QPM allele and three opaque RILs were homozygous for the W64Ao2 allele. Developing kernels were obtained from the selected RILs 30 days after pollination (DAP), frozen in liquid nitrogen, and stored at - 80 °C until use.
Starch composition
The K-TSTA and K-AMYL assays (Megazyme, Wicklow, Ireland) were used to determine total starch and amylose contents from developing and mature endosperm samples, respectively. Amylopectin content was obtained by difference.
Starch swelling power
It was determined according to Konik-Rose et al. (2001). About 40 mg of flour were mixed with 1 mL of water, vortexed, and then incubated for 30 min at 92.5 °C. The mixture was cooled and centrifuged (17,000 x g, 10 min). The swelling power was calculated by dividing the weight of the residue by the initial weight of flour.
Gelatinization enthalpy
Flour samples (2 mg) were mixed with deionized water (7 µL) and sealed in hermetic pans for analysis with a differential scanning calorimeter (DSC-2920, TA Instruments, New Castle, USA) calibrated with indium; an empty pan was used as a reference. The heating rate was 10°C/min over the temperature range (30-120 °C). Gelatinization enthalpy (∆HG) was calculated with TA Universal Analysis 2000 software (TA Instruments).
Genetic mapping
MapManager QTxb20 software (Manly et al., 2001) and SSR linkage groups previously reported for the RIL population (Holding et al., 2011) were used to identify QTL associated with starch composition, starch swelling, and gelatinization enthalpy by composite interval mapping (CIM) (Zeng, 1994). CIM was conducted with an additive regression model, and permutation tests (1000 shuffles) on individual chromosomes were performed to establish the significant threshold value of the likelihood ratio statistic (LRS) (Churchill and Doerge, 1994).
RNA isolation and cDNA synthesis
Total RNA was isolated from developing endosperm (30 DAP) according to Holding et al. (2008). Endosperms were ground in 200 µL of RNA extraction buffer [50 mmol/L Tris-HCl, pH 8, 150 mmol/L LiCl, 5 mmol/L EDTA, pH 8, 10 g/L SDS], added with 200 µL of phenol/chloroform (1:1) pH 8, mixed and placed 5 min on ice. The aqueous phase was separated (13,000 x g, 10 min, 4 °C). Starch was removed using 1 mL trizol reagent (Life Technologies), followed by the addition of 200 µL of phenolchloroform (1:1), and then centrifuged (10,000 x g, 10 min, 4 °C). Nucleic acids were precipitated with one volume of isopropanol, centrifuged (10,000 x g, 10 min, 4 °C), the pellet washed with 70 % ethanol and resuspended in 50 µL DEPC water. The obtained RNA was treated with DNase using the DNase I Kit (Sigma-Aldrich, St. Louis, USA). cDNA was synthesized from 2 µg of RNA using the Go Script TM reverse transcription system kit (Promega, Madison, USA).
Gene expression analysis
RT-qPCR was used to determine the 27-kDa γ-zein and starch biosynthesis genes transcript levels. The cDNA (75 ng) was amplified using the Power SYBR® Green PCR Master Mix (Applied Biosystems, UK). The PCR primers (Table 1) were designed using the Primer-BLAST program (http://www. ncbi.nlm.nih.gov/tools/primer-blast). A real-time system StepOnePlusTM (Applied Biosystems, Carlsbad, USA) was used under the following conditions: 95 °C for 10 min, then 40 cycles at 95 °C (15 s) and 60 °C (1 min) with a ramp rate of 2.2 °C/s. The gene encoding retinoblastoma-related protein 1 (RRB1) was used as a control, and the expression values were calculated relative to the parental line K0326Y QPM (vitre-ous), which is the donor of the modifier genes that convert the soft endosperm of W64Ao2 to a vitreous phenotype in the RILs. The relative expression was calculated according to the 2-∆∆CT method (Livak and Schmittgen, 2001) using the following equation: 2[(WX-WC) - (GX-GC)], where X= gene of interest, C= control gene (RRB1), W=average cycle threshold (Ct) of K0326Y QPM, and G= average Ct of the other genotypes.
Table 1 Gene-specific primers used for RT-qPCR.
| Gene | Primer | Amplicon size (bp) | |
|---|---|---|---|
| γ-zein | Fw | GTATAAGTCCTAAAGTGGTGAGGAACA1 | 240 |
| Rv | GAGCTGTAAGCATGTTATCAGATGAG1 | ||
| Bt2 | Fw | ATTACCGTTGCTGCCCTACC2 | 115 |
| Rv | ACTGCTCTCCTTTCGGTTTCTC2 | ||
| Wx1 | Fw | GAGGACGTCGTGTTCGTCTG3 | 245 |
| Rv | CAGTTGATCTTCCGGCCTTC3 | ||
| Ae1 | Fw | CCGACGCTGGACTATTTGGT3 | 165 |
| Rv | GCAACGAGTACCCCGCTATC3 | ||
| Zpu1 | Fw | GCTTCAGTTGCATCCAGTGC3 | 250 |
| Rv | TGGGCAACGTCTTATTCAATC3 | ||
| RRB1 | Fw | GCTGTTTCTGGTTATGTCTGTCCT3 | 138 |
| Rv | CTTTTGAGTACTTCTGTGCCTGAC3 |
γ-zein: 27kDa γ-zein; Bt2: Brittle-2 (ADP-glucose pyrophosphorylase, small subunit); Wx1: Waxy-1 (Granule-bound starch synthase 1); Ae1: Amylose extender 1 (Starch-branching enzyme); Zpu1: Pullulanase-type starch debranching enzyme 1; RRB1: Retinoblastoma-related protein 1; Fw, Forward; Rv, Reverse. 1Holding et al. (2011); 2Huang et al. (2011); 3Jia et al. (2013).
Zein Extraction, Quantification, and SDS-PAGE Analysis
Developing endosperms were lyophilized and ground into a fine powder to obtain the zein fractions as described by Wallace et al. (1990). Total protein from 50 mg of endosperm flour was extracted overnight with 1 mL of borate buffer (12.5 mmol/L Na2B4O7-10H2O, 10 g/L SDS, 20 mL/L 2-mercaptoethanol, pH 10) at 37 °C. The extract was adjusted with ethanol (700 mL/L) to precipitate the non-zein fraction. Zein protein extracts (equivalent to 15 mg of flour) were vacuum-dried and resuspended in 100 µL of deionized water. Five microliters were separated in 0.15 mg/mL polyacrylamide gels (SDS-PAGE) and stained with Coomassie Blue R250. The Quantity One software (BioRad, Hercules, CA) was used to quantify by densitometry the protein bands.
Isolation of starch granules, extraction and identification of starch granule-associated proteins (SGAP)
Starch granules were isolated from developing (30 DAP) kernels, according to Grimaud et al. (2008). Kernels were soaked in 3 g/L sodium metabisulfite and 850 mL/L lactic acid (pH 3.8) for 48 h at 50 °C, and then the germ and pericarp were manually dissected. Endosperms were ground with a mortar and pestle in starch extraction buffer [50 mmol/L Tris-HCl, pH 7, 100 mL/L glycerol, 10 mmol/L EDTA, 1.25 mmol/L DTT] at 4 °C, passed through a 100 µm filter and centrifuged at 13,000 x g for 15 min. Starch was washed five times with cold extraction buffer, 950 mL/L ethanol, and acetone and then dried for 12 h. For SGAP extraction, about 150 mg of starch were washed at 4 °C with water, gelatinization buffer [62.5 mmol/L Tris-HCl, pH 6.8, 20 g/L SDS, 50 mL/L β-mercaptoethanol], and 950 mL/L ethanol to remove superficial proteins. Then, starch samples were incubated with gelatinization buffer for 15 min in a boiling water bath to release SGAP. The paste formed was frozen for 1 h, thawed for 20 min at 40 °C in a water bath and then centrifuged (13,000 x g, 30 min). The supernatant was collected and mixed with an equal volume of 300 g/L TCA/Acetone for at least 2 h to precipitate proteins. After centrifugation for 15 min at 4 °C, the SGAP pellet was washed three times with cold acetone and dried overnight at room temperature. SGAP were resuspended in thiourea buffer [7 mol/L Urea, 2 mol/L Thiourea, 40 g/L CHAPS, 20 mmol/L DTT, 2 mL/LTriton-X100]. Protein extracts equivalent to 8 and 21 mg of starch were used for granule-bound starch synthase and low abundant SGAP, respectively; they were separated by SDS-PAGE using a 0.10 mg/mL polyacrylamide gel and identified by mass spectrometry according to Salazar-Salas et al. (2014).
Statistical analysis
Data correspond to the mean of three biological replicates with three technical replicates. It was analyzed using one-way ANOVA, and the Fisher test (p < 0.05) was used for mean comparisons. These analyses were performed using the software STATGRAPHIC plus version 5.1 (Statistical Graphics Corporation™, Maryland, USA).
Results and discussion
Identification of QTL associated with starch properties
The phenotypic variation of starch, amylose, amylopectin content, swelling power, and gelatinization enthalpy was determined in 103 RIL derived from K0326Y QPM and W64Ao2 (Table 2). A continuous variation was observed in the RIL population for every trait studied, and the extreme values were outside of those from the parental lines, indicating transgressive segregation. Composite interval mapping identified 26 QTL, which explained 66% of the total variation for the starch properties (Table 3). For total starch, five loci were identified on chromosomes 1 (qST-1), 4 (qST-4), 5 (qST-5), 7 (qST-7), and 10 (qST-10); the alleles contributing to this trait were mainly from K0326Y QPM, except for two loci contributed by the W64Ao2 alleles. Similar chromosomal locations associated with QTLs for starch content have been reported in different maize germplasm (Séne et al., 2001; Zhang et al., 2008; Dong et al., 2015).
Tabla 2 Variabilidad de propiedades fisicoquímicas de almidón en endo-spermo maduro de K0326Y QPM, W64Ao2 y líneas recombinantes puras (RIL) derivadas de su cruza.
Table 2 Variability of starch physicochemical properties in mature endos-perm of K0326Y QPM, W64Ao2 and recombinant inbred lines (RIL) derived from their cross.
| Property | K0326Y QPM | W64Ao2 | RILs |
|---|---|---|---|
| Total starch (mg /100 mg flour) | 74.00 ± 0.80a | 75.00 ± 0.80a | 63.21 - 76.79 |
| Amylose (mg /100 mg starch) | 25.54 ± 1.06a | 21.79 ± 0.49b | 15.56 - 32.29 |
| Amylopectin (mg /100 mg starch) | 74.46 ±1.06b | 78.21 ± 0.49a | 67.71 - 89.91 |
| Swelling power (g /g flour) | 7.19 ± 0.01b | 9.22 ± 0.08a | 5.89 - 10.09 |
| Gelatinization enthalpy (J/g) | 10.35 ± 0.13b | 11.54 ± 0.52a | 8.14 - 13.41 |
Different superscript letters between the parental lines K0326Y QPM and W64Ao2 indicate significant differences based on the Fisher test (P<0.05).
Table 3 Characteristics of QTLs associated with starch properties.
| Trait | QTL | Bin | Left-Right markers | LRS | Variance | Effect* |
|---|---|---|---|---|---|---|
| Total starch | qST-1 | 1.07-1.08 | Umc1278-Dupssr12 | 12.2 | 11 | -0.96 |
| qST-4 | 4.04-4.05 | Mmc0471-Bnlg1937 | 13.8 | 14 | 1.02 | |
| qST-5 | 5.03-5.04 | Umc1056-Bnlg2323 | 12.7 | 12 | -0.96 | |
| qST-7 | 7.04 | Umc1029-Umc1944 | 12.2 | 11 | 0.86 | |
| qST-10 | 10.04-10.06 | Umc1053-Umc2190 | 12.7 | 11 | -0.99 | |
| Amylose | qAM-1 | 1.05-1.06 | Umc1076-Umc1335 | 11.3 | 9 | -1.14 |
| qAM-3 | 3.05-3.06 | Umc1167-Bnlg197 | 15.6 | 14 | -1.32 | |
| qAM-5 | 5.04-5.05 | Bnlg2323-Mmc0081 | 13.0 | 12 | -1.29 | |
| qAM-7 | 7.02 | Umc1036-γ zein | 19.6 | 19 | -1.65 | |
| qAM-9 | 9.03-9.04 | Bnlg127-Umc1771 | 16.4 | 14 | -1.27 | |
| Amylopectin | qAP-1 | 1.05-1.06 | Umc1076-Umc1335 | 11.3 | 9 | 1.14 |
| qAP-3 | 3.05-3.06 | Umc1167-Bnlg197 | 15.6 | 14 | 1.32 | |
| qAP-5 | 5.04-5.05 | Bnlg2323-Mmc0081 | 13.0 | 12 | 1.29 | |
| qAP-7 | 7.02 | Umc1036-γ zein | 19.6 | 19 | 1.65 | |
| qAP-9 | 9.03-9.04 | Bnlg127-Umc1771 | 16.4 | 14 | 1.27 | |
| SP | qSP-1 | 1.05-1.06 | Umc1076-Umc1335 | 12.8 | 12 | 0.30 |
| qSP-2 | 2.08-2.09 | Umc1516-Umc1551 | 11.7 | 10 | 0.28 | |
| qSP-5 | 5.05 | Umc1264-Bnlg278 | 11.9 | 10 | 0.31 | |
| qSP-7 | 7.02 | Umc1036-γ zein | 23.7 | 12 | 0.39 | |
| qSP-9 | 9.03 | Wx1-Bnlg127 | 28.5 | 23 | 0.43 | |
| ∆HG | qGE-1.1 | 1.03 | Bnlg176-Bnlg439 | 19.3 | 16 | 0.42 |
| qGE-1.2 | 1.05-1.06 | Umc1076-Umc1335 | 10.7 | 9 | 0.41 | |
| qGE-2 | 2.08-2.09 | Umc1516-Umc1551 | 13.6 | 12 | 0.41 | |
| qGE-5 | 5.08-5.09 | Umc1225-Umc1153 | 11.0 | 10 | 0.36 | |
| qGE-7 | 7.02 | Umc1036-γ zein | 11.8 | 11 | 0.40 | |
| qGE-9 | 9.03 | Wx1-Bnlg127 | 14.9 | 13 | 0.47 |
LRS, likelihood ratio statistic. *The additive effect is negative for the K0326Y-QPM allele and positive for the W64Ao2 allele. SP, Swelling power. ∆HG, Gelatini-zation enthalpy.
Five QTLs were identified for amylose content (Table 3), and the favorable alleles came from K0326Y QPM. These loci were also identified for amylopectin content but in this case, the positive additive effect corresponded to alleles from W64Ao2 (high amylopectin). The major QTLs for these traits were in bins 3.05-3.06 (qAM-3, qAP-3), 7.02 (qAM-7, qAP-7), and 9.03-9.04 (qAM-9, qAP-9), explaining 47 % of the observed variation. The loci on chromosomes 1, 7, and 9 coincided with the QTL reported by Holding et al. (2011) for traits associated with o2 endosperm modification (vitreousness, density, and hardness) in the same RILs. A significant correlation was observed between amylose content and vitreousness in the RIL population (r = 0.52, p < 0.01).
Five QTL were identified for starch swelling power (SP) on chromosomes 1 (qSP-1), 2 (qSP-2), 5 (qSP-5), 7 (qSP7), and 9 (qSP-9) (Table 3), highlighting qSP-9 that explains 23 % of the variation. The qSP-1, qSP-7, and qSP-9 loci coincided with those identified for amylose (Table 3) and vitreousness (Holding et al., 2011). Alleles from W64Ao2 (soft endosperm) contributed to the variation, which corresponds with the high SP and low amylose values of this parental line and the inverse correlation observed for these traits in the RILs (r = -0.413, p < 0.01). SP is frequently used as an indirect measure of water uptake during starch gelatinization (Konik-Rose et al., 2001) and is influenced by the granule composition. The asymmetric distribution of amylopectin molecules favors the entry of water and thereby the swelling power, whereas amylose can form amylose -lipids complexes that inhibit swelling (Tester and Morrison, 1990).
Four of the six QTL identified for gelatinization enthal-py were coincident with those for SP on chromosomes 1 (qGE-1.2), 2 (qGE-2), 7 (qGE-7), and 9 (qGE-9), which together explained 51 % of the ∆HG variation (Table 3). Similar to SP, loci on chromosomes 7 and 9 were coincident with those previously reported for amylose (Table 3) and vitreousness (Holding et al., 2011); the W64Ao2 alleles contributed to the ∆HG variation, which corresponded with the higher gelatinization enthalpy observed for this parental line (Table 2). Gelatinization enthalpy has been used as a measure of starch crystallinity, which is related to the content of amylopectin (Matveev et al., 2001).
Candidate genes for the QTLs were proposed based on their proximity to flanking markers in the Maize Genome Database (https://www.maizegdb.org); colocations between candidate genes and the QTLs were considered significant when the distances were lower than 10-20 cM (Séne et al., 2001). The flanking markers of QTLs for starch content and composition on bins 4.05 (Bnlg1937) and 5.04 (Bnlg2323) were found near Brittle -2 (Bt2) and Amylose extender-1 (Ae1) genes, respectively. Bt2 encodes for the small subunit of ADP glucose pyrophosphorylase and Ae1 for starch branching enzyme IIb (SBEII). The QTL on bin 9.03 (Wx1-Bnlg127) corresponds to the waxy gene (Wx1), which encodes for the GBSSI enzyme, responsible for the synthesis of amylose in the starch granule (Denyer et al., 2001).
The QTL on chromosome 7 (Umc1036-γ zein) was located in a locus corresponding to the 27-kDa γ-zein, which was previously reported for vitreousness, hardness, and density in the same RIL population (Holding et al., 2011). Transcription factors that regulate zein genes also appear to control starch synthesis (Zhang et al., 2016). A QTL on chromosome 1 was observed for all traits, but its effect is unclear since no candidate genes affecting starch properties were identified close to this locus; however, the flanking marker in bin 1.05 (Umc1076) is close to a gene for 19 kDa α-zein (az19D1). Salazar-Salas et al. (2014) reported higher levels of 19 kDa α-zein in vitreous than opaque lines and a highly significant correlation (r = 0.75, p < 0.01) between the contents of this protein and those of 27 kDa γ-zein in the RIL population derived from K0326Y QPM and W64Ao2. Both proteins may be contributing to protein bodies formation and the vitreous phenotype.
Starch composition in developing endosperm of QPM lines contrasting in vitreousness
The starch composition was determined in developing endosperms (30 DAP) of K0326Y QPM, W64Ao2, and six RIL contrasting in kernel vitreousness (Table 4) to investigate its relationship with the expression of starch biosynthesis genes. The starch composition of mature endosperms (50 DAP) was included as a reference. At 30 DAP, total starch ranged from 59.1 to 64.6 %; these values were lower than the 88.9 % reported by Li et al. (2007) in B73 maize at the same developmental stage, which could be attributed to differences in genotype, growing conditions and method used for starch isolation. No significant differences were found in total starch content between the parental lines K0326Y QPM and W64Ao2. Pereira et al. (2008) reported a similar behavior between an opaque-2 and a QPM line at 30 DAP. The starch content increased to an average of 75 % in mature endo-sperms (Table 4).
Tabla 4 Composición de almidón en endospermo en desarrollo (30 DDP) y maduro (50 DDP) de K0326Y QPM, W64Ao2, y líneas recombinantes puras seleccionadas contrastantes en el genotipo de marcadores que flanquean loci asociados con la modificación del endospermo en QPM.
Table 4 Starch composition in developing (30 DAP) and mature (50 DAP) endosperms of K0326Y QPM, W64Ao2, and selected recombinant inbred lines (RILs) contrasting in the genotype of markers flanking loci associated with endosperm modification in QPM.
| Maize line | Starch* | Amylose** | Amylopectin** | ||||
| 50 DAP | 30 DAP | 50 DAP | 30 DAP | 50 DAP | 30 DAP | ||
| K0326Y-QPM | V | 59.4 ± 1.5d | 74.0 ± 0.8c | 20.0 ± 0.7a | 25.5 ± 1.1a | 80.0 ± 0.7c | 74.5± 1.1e |
| W64Ao2 | O | 59.6 ± 1.1cd | 75.0 ± 0.8bc | 17.2 ± 0.5b | 21.8 ± 0.5b | 82.8 ± 0.5b | 78.2 ± 0.5d |
| 33 | V | 61.5 ± 1.4bc | 77.6 ± 1.2a | 16.5 ± 0.4b | 21.7 ± 0.4bc | 83.5 ± 0.4b | 78.3 ± 0.4cd |
| 338 | V | 61.0 ± 1.4bcd | 75.8 ± 1.7b | 16.6 ± 0.7b | 22.0 ± 0.3b | 83.4 ± 0.7b | 78.0 ± 0.3d |
| 346 | V | 62.3 ± 1.3b | 75.9 ± 0.6ab | 17.1 ± 0.4b | 21.9 ± 0.5b | 82.9 ± 0.4b | 78.0 ± 0.5d |
| 55 | O | 64.6 ± 0.7a | 75.7 ± 0.7b | 14.2 ± 0.4c | 19.9 ± 0.7de | 85.8 ± 0.4a | 80.1 ± 0.7ab |
| 73 | O | 62.5 ± 1.3ab | 74.9 ± 0.6bc | 14.6 ± 0.7c | 19.5 ± 0.4e | 85.3 ± 0.7a | 80.5± 0.4a |
| 185 | O | 59.5 ± 1.1d | 76.1 ± 0.6ab | 16.8 ± 0.1b | 20.7 ± 0.5cd | 83.2 ± 0.1b | 79.3 ± 0.5bc |
DAP, Days after pollination; V, vitreous; O, opaque; * Total starch expressed as mg per 100 mg of endosperm flour; ** Amylose and amylopectin expressed as mg per 100 mg of starch. For each property and developmental stage different superscript letters in the same column indicate significant differences between the maize lines based on the Fisher test (P < 0.5).
The amylose content of developing endosperm from K0326Y QPM (20.0 %) was significantly higher (p ≤ 0.05) than that of W64Ao2 (17.3 %), which corresponded with the higher amylose content observed in the vitreous RIL with respect to the opaque lines (Table 4). The same behavior was observed in the mature endosperm. These results agree with those of Dombrink-Kurtzman and Knutson (1997) and Gayral et al. (2015), who observed higher amylose levels in starch from the vitreous region of maize endosperm with respect to starch from the floury area.
Expression analysis of QTL candidate genes
The molecular basis of QTLs associated with starch properties was investigated by measuring the expression of a 27-kDa γ-zein gene (Fig. 1) and four genes related to starch biosynthesis (Bt2, Wx1, Ae1, and Zpu1) (Fig. 2) in the parental lines and six RILs contrasting in vitreousness and the genotype of flanking markers. A late middle stage (30 DAP) of seed development was chosen because the endosperm cells have high levels of gene expression and large amounts of starch and storage proteins that may be associated with the vitreous phenotype in mature endosperm. The relative expression of the 27-kDa γ-zein gene was significantly higher in K0326Y QPM and the vitreous RIL with respect to W64Ao2 and the opaque lines (Fig. 1A). Holding et al. (2011) also observed a higher expression of this gene in vitreous RILs after 18 DAP, indicating very low levels of transcripts for opaque samples as observed in the present study. This expression pattern was confirmed at the protein level in developing (30 DAP) endosperms (Fig. 1B); the mean value of 27-kDa γ-zein was 21.8 % higher in vitreous than opaque RILs. Sethi et al. (2021) also reported that the prolamin-like fraction that includes 27-kDa γ-zein showed higher accumulation in QPM lines compared to opaque-2 genotypes after 30 and 45 DAP. Salazar-Salas et al. (2014) reported a significant correlation (r = 0.58, p < 0.01) between the levels of 27-kDa γ-zein and vitreousness in 283 RIL derived from the cross between K0326Y QPM and W64Ao2. These results support the essential role of γ-zein in the vitreous phenotype of Quality Protein Maize (Wu et al., 2010), whose enhanced expression was associated with a duplication at the 27-kDa γ-zein locus (Liu et al., 2016).
Figura 1 Expresión relativa del gen de γ-zeína y perfil de zeínas en endospermo en desarrollo (30 DDP) de líneas contrastantes en vitrosidad y el genotipo de marcadores que flanquean QTLs para esta característica. (A) Expresión relativa del gen de γ-zeína de 27 kDa. Letras diferentes indican diferencias significativas entre las líneas de maíz basado en la prueba de Fisher (P<0.05). (B) Separación por SDS-PAGE de zeínas de endospermo en desarrollo (30 DDP) (B). Los marcadores de peso molecular (kDa) se muestran en la izquierda y en la derecha se indican las principales sub-fracciones de zeínas.
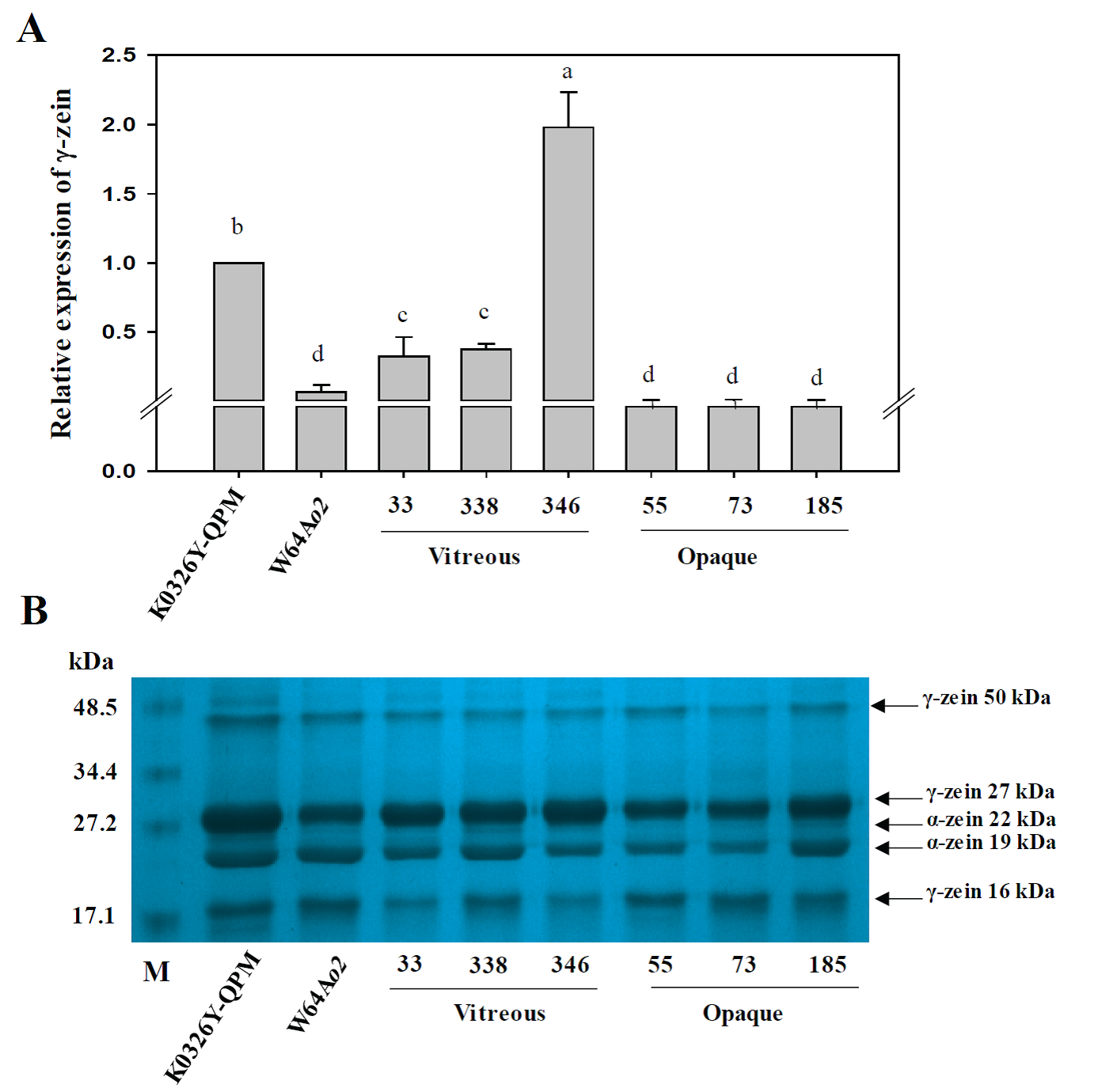
Figure 1 Relative expression of γ-zein gene and zein profiles in developing endosperm (30 DAP) of lines contrasting in vitreousness and the genotype of QTLs-flanking markers for this trait. (A) Relative expression of the 27-kDa γ-zein gene. Different letters indicate significant differences between the maize lines based on the Fisher test (P<0.05). (B) SDS-PAGE separation of zein proteins. Molecular weight (kDa) standards are shown on the left, and the main zein sub-fractions are indicated on the right.
Figura 2 Expresión relativa de genes de síntesis de almidón en endospermo en desarrollo (30 DDP) de líneas de maíz contrastantes en vitrosidad y el genotipo de marcadores que flanquean QTLs para esta característica. El gen de la proteína relacionada a retinoblastoma 1 (RRB1) fue usado como referencia y los valores de expresión son relativos al de la línea K0326Y QPM. Letras diferentes indican diferencias significativas entre las líneas de maíz basado en la prueba de Fisher (P < 0.05).
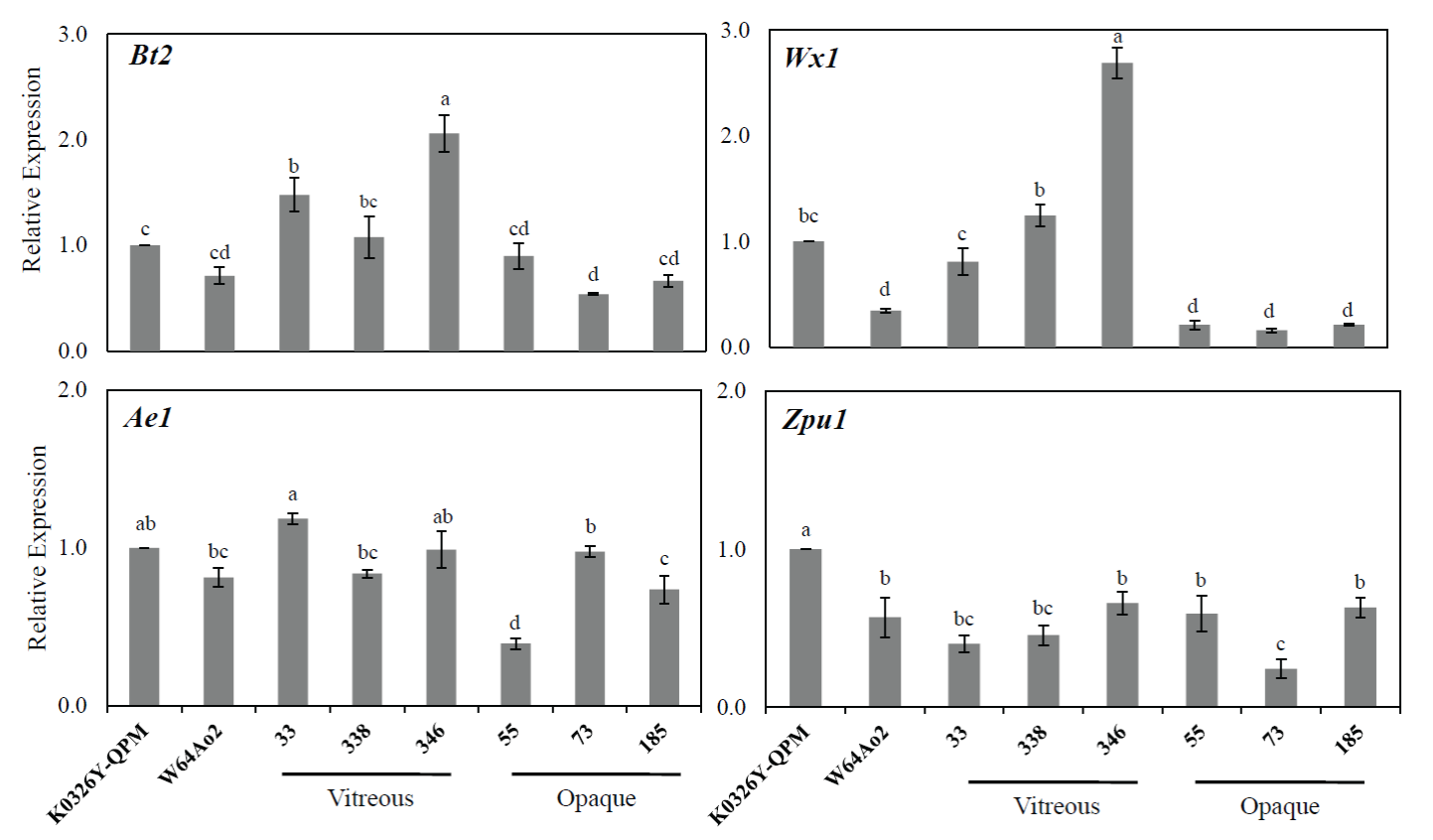
Figure 2 Relative expression of starch biosynthesis genes in developing endosperm (30 DAP) of maize lines contrasting in vitreousness and the genotype of QTLs-flanking markers for this trait. The retinoblastoma-related protein-1 (RRB1) gene was used as a reference, and the expression values are relative to that of the K0326Y QPM line. Different letters indicate significant differences between the maize lines based on the Fisher test (P < 0.05).
The Bt2 gene encodes for ADP-glucose pyrophos-phorylase, a key enzyme in starch biosynthesis (Giroux and Hannah, 1994). The relative expression of this gene was not significantly different between the parental lines K0326Y QPM and W64Ao2 (Fig 2), which corresponded with the similar starch content observed in developing endosperm from these lines (Table 4). There were some differences in Bt2 expression between the RILs, where the vitreous lines gen-erally showed higher transcript levels than the opaque lines (Fig. 2). The over-expression of Bt2 (~2.5-fold) was associated with a 7.5 % increase in starch content in developing (30 DAP) maize endosperm (Li et al., 2011). However, the higher transcript levels of Bt2 observed in some vitreous lines were not associated with significant changes in starch content (Table 4). The Wx1 gene maps close to the QTL for amylose and amylopectin on chromosome 9 (Bnlg127). Wx1 encodes for granule -bound starch synthase I (GBSSI), the enzyme responsible for amylose synthesis (Denyer et al., 2001). The Wx1 expression was three-fold greater in K0326Y QPM than W64Ao2 and eight-fold higher in vitreous than opaque RILs (Fig. 2). These results are in agreement with the higher amy-lose content observed in K0326Y QPM and the vitreous RIL with respect to W64Ao2 and the opaque RILs (Table 4). The increased expression of Wx1 may lead to a more significant accumulation and activity of GBBSI that results in the high amylose content associated with the vitreous phenotype.
The Ae1 gene encodes for the starch branching en-zyme IIb (SBEIIb), which catalyzes the formation of the α-1,6 glycosidic bonds during the synthesis of amylopectin (Nishi et al., 2001). Although the amylopectin content, in general, was higher in W64Ao2 and opaque RILs than K0326Y QPM and vitreous RILs (Table 4); there were no significant differ-ences in Ae1 expression between opaque and vitreous lines at 30 DAP (Fig. 2). However, the expression of this gene was higher in the opaque lines at an earlier stage of endosperm development (20 DAP) (data not shown) and may explain the higher amylopectin content of these lines. Jia et al. (2013) also found a higher expression of Ae1 in W64Ao2 with respect to its normal counterpart W64A+ at 22 DAP, which could favor the production of starch granules with larger crystalline re-gions associated with the opaque phenotype.
The Zpu1 gene encodes the pullulanase-type starch debranching enzyme, which hydrolyzes the α-1,6 glycosidic bonds of pullulan, a linear polymer of maltotriose units pro-duced during starch metabolism (Nakamura et al., 1996). The expression level of this gene in the endosperm of the paren-tal line W64Ao2 was half of that in K0326Y QPM (Fig. 2), but there were no significant differences between the opaque and vitreous RILs.
Accumulation of starch granule-associated proteins
Starch granule-associated proteins (SGAP) from developing and mature endosperms (Fig. 3, Table 5) were analyzed because the GBSSI-encoding gene (Wx1) was found to be differentially expressed between the vitreous and opaque lines. The most abundant protein (~55 kDa) was GBSSI and showed more significant accumulation in deve-loping endosperms from the vitreous lines (Fig. 3), which corresponds with the higher amylose content observed in these samples (Table 4). The other SGAP corresponded to the enzymes starch synthase I (SSI), starch synthase IIa (SSIIa), starch branching IIb (SBEIIb), sucrose synthase 1 (SH1), and pyruvate phosphate dikinase 2 (PPDK2), which are involved in amylopectin synthesis and starch metabolism. Unfortuna-tely, the low abundance of these proteins did not allow to register differences in accumulation between the vitreous and opaque lines. Other authors previously reported similar SGAP patterns in maize (Grimaud et al., 2008; Juárez-García et al., 2013; Salazar-Salas et al., 2014). PPDK and SH1 form complexes with some starch biosynthetic enzymes like SSIII, SSIIa, SBEIIa, and SBEIIb in maize amyloplasts and they may regulate the carbon partitioning between metabolic pathways (Hennen-Bierwagen et al., 2009).
Figura 3 Separación por SDS-PAGE de proteínas asociadas al gránulo de almidón en endospermo en desarrollo (30 DDP) de líneas contrastantes en vitrosidad y el genotipo de marcadores que flanquean QTL asociados con esta característica. Los marcadores de peso molecular (kDa) se indican a la izquierda y la identidad de las proteínas se indica a la derecha. Las proteínas fueron identificadas por espectrometría de masas (Tabla 5). GBSSI, Almidón sintasa unida al gránulo I; SSI, Almidón sintasa I; SSIIa, Almidón s intasa IIa; SBEIIb, Enzima ramificadora de almidón IIb; SHI, Sacarosa sintasa I; PPDK2, Piruvato fosfato dicinasa 2.
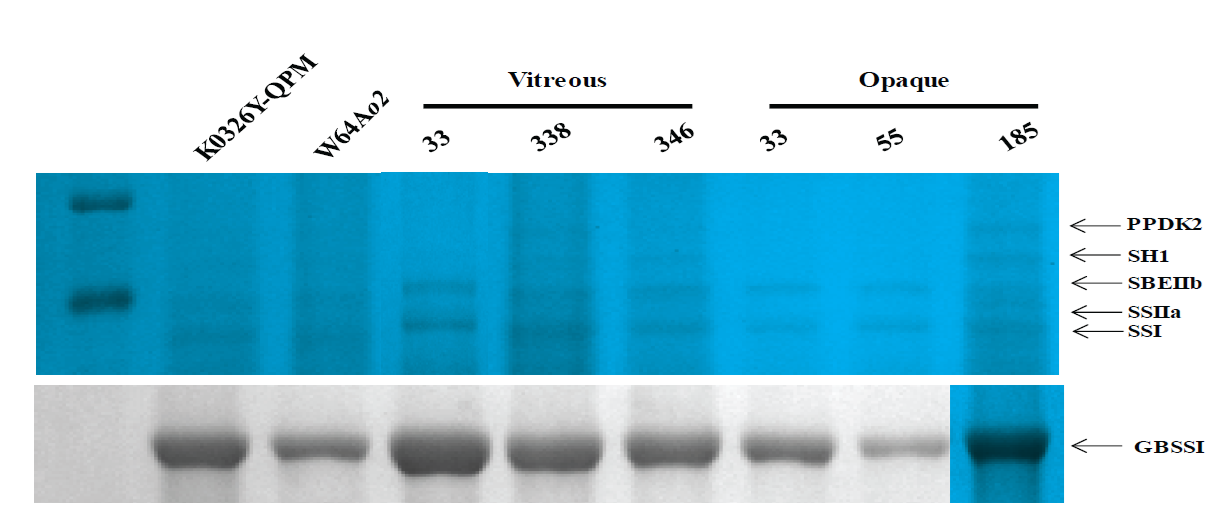
Figure 3 SDS-PAGE analysis of starch granule-associated proteins from developing endosperm (30 DAP) of lines contrasting in vitreousness and the genotype of markers flanking QTLs for this trait. Molecular weight (kDa) standards are shown on the left, and the corresponding proteins are indicated on the right. Protein bands were identified by mass spectrometry (Table 5). GBSSI, Granule-bound starch synthase I; SSI, Starch synthase I; SSIIa, Starch synthase IIa; SBEIIb, Starch branching enzyme IIb; SH1 sucrose synthase 1; PPDK2, Pyruvate phosphate dikinase 2.
Table 5 Identification by tandem mass spectrometry of starch granule associated proteins.
| Accession | Theoretical | Observed | Sequence | ||
|---|---|---|---|---|---|
| No. | Protein name | Mw (kDa) | Mw (kDa) | Coverage (%) | Peptides matched |
| A9Y0P1 | Granule-bound | EALQAEVGLPVDR | |||
| starch synthase | 65.8 | 63.2 | 21.52 | FSVDCNVVEPADVK | |
| [Zea mays] | FEPCGLIQLQGMR | ||||
| O49064 | Starch synthase I [Zea mays] | 70.5 | 75.6 | 12.3 | GAVVTADRIVTVSK EHKSSWEGLMK GIDLIQLIIPDLMR |
| Q6RG16 | Starch synthase IIa, [Zea mays] | 79.9 | 81.7 | 9.33 | ITAGADVLVMPSR MLQHLEREHPNK APRDAALVRAEAEAGGK |
| O81387 | Starch branching enzyme IIb [Zea mays] | 90.6 | 89.6 | 17.9 | VVLDSDAGLFGGFSR KAVMVPEGENDGLASR FIPGNNNSYDKCRR |
| K7V5Z8 | Shrunken1 [Zea mays] | 91.7 | 96.0 | 10.27 | ELANLVIVAGDHGK LLPDAAGTTCGQR VIGTEHTDIIR |
| K7UZT6 | Pyruvate, phosphate dikinase 2 [Zea mays] | 105.6 | 105.6 | 21.63 | EGEWLSLNGSTGEVILGK IAVDMVNEGLVER AWTAFPGSAQEGIGR |
Conclusions
The endosperm modification in QPM is associated with changes in the expression of starch biosynthesis genes, such as Wx1 that showed higher transcript levels in the endosperm of the vitreous lines (K0326Y QPM, 33, 338, 346), which resulted in a more significant accumulation of GBSSI and consequently the synthesis of starch with a higher proportion of amylose during kernel development. As a result, starch granules with more amorphous regions may favor their packing and association, contributing to the vitreous phenotype. The information about the mechanisms involved in the modification of the opaque-2 endosperm will facilitate the development of QPM genotypes with better agronomic and processing characteristics.











 nova página do texto(beta)
nova página do texto(beta)


