Introduction
In December 2019, a series of atypical pneumonia cases occurred in Wuhan, Hubei province, China, caused by a new b-coronavirus named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by the International Committee on Taxonomy of Viruses (ICTV)1-3.
Coronaviruses are highly diverse RNA viruses of the Coronaviridae family divided into four genera: alpha, beta, gamma, and delta. They are widely distributed in humans and other mammals. These viruses cause different illnesses: from common flu in its less aggressive form to pneumonia; in some cases, they cause severe acute respiratory syndrome (SARS) that requires ventilatory or mechanical support, and in the most severe cases, they cause death2-5.
The World Health Organization (WHO) declared a global pandemic on March 11, 2020, as part of a global effort to coordinate the management of the impact caused by the SARS-CoV-2 virus. This pandemic has caused 462,684 confirmed cases (81,968 cases in China, including 11,977 severe cases) and 20,834 deaths (3293 deaths in China), figures up to March 26, 2020. Some studies have suggested that it could take more than a decade for the world to recover socially and economically2,3.
According to the WHO, the implementation of systems to diagnose cases, timely identification of suspected cases, collection and delivery of samples to reference laboratories, and the implementation of molecular detection protocols for SARS-CoV-2, depending on the capacity of each laboratory, are among the priority strategies to be followed for pandemic management. Therefore, the coordination and collaboration of the clinical laboratory to support diagnosis based on molecular tests is of significant importance for the timely care of patients and decision-making in the healthcare sector2-5.
Clinical and epidemiological studies have shown that patients with COVID-19 present symptoms ranging from mild to severe, including fever, fatigue, severe pneumonia, acute respiratory distress syndrome (ARDS), severe inflammatory response syndrome (SIRS), target organ damage, and multiple organ failure (MOF). According to different studies, SARS-CoV-2 infection is associated with significant morbidity and mortality ranging from 4.3% to 15%, especially in patients with chronic medical conditions. At least one in five cases requires intervention in intensive care units, which are limited in developing countries1-5.
Recently, some laboratory parameters useful to predict the course of the disease have been described: some clinical features include peripheral blood leukocyte count (TCD4+ and TCD8+ cells) decrease, and the increase of proinflammatory cytokines in serum such as interleukin 6 (IL-6) and, interferon-gamma (IFN-g). Moreover, acute-phase proteins (C-reactive protein, fibrinogen, ferritin) and cardiac biomarkers (troponin and BNP) quantification, the assessment of thrombosis risk, and liver and renal function evaluation tests are essential elements for the clinician to establish therapeutic and prophylactic measures when treating patients with COVID-195-7.
Several studies have reported that child infection represents from 1-5% of the total cases. This population often shows milder symptoms than the adult population, and deaths are extremely rare5-7.
This review aimed to provide information regarding the implementation of molecular tests to detect SARS-CoV-2 in the Hospital Infantil de México Federico Gómez (HIMFG) clinical laboratory. These procedures are based on the molecular diagnostic protocols established by the WHO Laboratory testing for 2019 novel coronavirus (2019-nCoV) in suspected human cases.
Molecular diagnosis of COVID-19
SARS-CoV-2 is a single-stranded RNA virus with a 29,903 nucleotides genome, making it the second-largest known RNA genome. It consists of two untranslated regions (UTR) at the 5 and 3 ends and 11 open reading frames (ORFs), which encode 27 proteins6-10.
SARS-CoV-2 confirmation is based on detecting its RNA by real-time reverse transcription-polymerase chain reaction (RT-PCR) assays. In general, SARS-CoV-2 identification is carried out to detect three genes: N, E, and RdRp genes (when using the Berlin protocol). Both the E and N genes serve as screening genes to detect any b-coronavirus associated with bats, while the RdRp gene is specific for SARS and SARS-like coronaviruses (including 2019-nCoV)10-18.
Tests for SARS-CoV-2 identification should be considered for those patients who meet the case definition. If possible, it is recommended to use the laboratory algorithm for influenza detection suggested by PAHO for influenza surveillance.
Additionally, sequencing platforms can be used for virus identification in laboratories with both Sanger and Next-Generation sequencing technology12-16.
Although serological methods based on the detection of IgM and IgG immunoglobulin antibodies exist in the market, they are not recommended, mainly because the dynamics between the response and production of antibodies during the infection are not entirely established; also, cross-reactivity with other coronaviruses makes the interpretation of the results difficult. Molecular detection of SARS-CoV-2 by RT-PCR has higher specificity and sensitivity (95-97%, confidence interval (CI): 83-97%); therefore, a positive result confirms the detection of the virus6-14.
The efficiency of RT-PCR in microbiology has been described in multiple diagnostic algorithms, and now it is widely used in diagnostic virology. As a first step, the reverse transcriptase enzyme is used to synthesize complementary DNA (cDNA) from RNA obtained from SARS-CoV-2 present in nasopharyngeal swab samples. Subsequently, the region of interest of the cDNA is amplified with the use of specific primers.
Early and timely detection of SARS-CoV-2 cases allows the implementation of isolation measures to reduce and delay the pandemic peak, allowing greater responsiveness for the healthcare sector. Molecular detection tests for SARS-CoV-2 are critical for diagnosing the disease, understanding its epidemiology, and managing the cases, leading to a decrease in infections8-10.
In our country, the Epidemiological Diagnostic and Reference Institute (InDRE, for its Spanish acronym) has developed a protocol for SARS-CoV-2 molecular identification, which has been transferred to the national network of public health laboratories, national health institutes, and other institutions that generate information for epidemiological surveillance.
It is essential to follow the general laboratory guidelines established by the InDRE and those documented in the Norma Oficial Mexicana (Mexican Official Standard) NOM-017-SSA2-2012 for epidemiological surveillance.
Currently, in our country, commercial tests must be subjected to an evaluation process by the InDRE for the accreditation of their analytical performance (Table 1). The complete list is available at the following link: https://www.gob.mx/cms/uploads/attachment/file/561223/Listado_de_estuches_comerciales_utiles_para_el_diagn_stico_de_SARS-CoV-2.pdf
Table 1 Description of molecular tests for the diagnosis of SARS-CoV-2 during the COVID-19 contingency approved by the InDRE in Mexico
| Name of the test | Catalog number | Manufacturer | Analytical sensitivity (detection limit) | Analytical specificity (absence of cross-reactivity) | Genes detected | Number of reactions per sample |
|---|---|---|---|---|---|---|
| Berlin reference test performed at InDRE and the National Network of Public Health Laboratories (Red Nacional De Laboratorios de Salud Pública) | Not available commercially | Institutional implementation (InDRE) | 10 copies/reaction | 100% | E, RdRp, RNAse P* | 3 |
| Novel Coronavirus (2019-nCoV) Nucleic Acid Diagnostic Kit (PCR-Fluorescence Probing) | S3102E | Sansure Biotech Inc. | 4 copies/reaction | 100% | ORF1ab, N, RNAse P* | 1 |
| MOLgen SARS-CoV-2 Real Time RT-PCR Kit | MESARS-CoV-2 | Adaltis S. r. l. | 100 copies/reaction | 100% | RdRp, N, E | 1 |
| QuantiVirusTM SARS-CoV-2 Test Kit | DC-11-008 | DiaCarta, Inc. | 50 copies/reaction | 100% | ORF1ab, N, E, RNAse P* | 4 |
| SARS-CoV-2 Nucleic Acid Detection Kit (PCR-Fluorescent Probe Method) | CoV2-32 | Zybio Inc. | 100 copies/reaction | 100% | ORF1ab, N | 1 |
| BioFire® COVID-19 Test | 423744 | BioFire Defense, LLC | ** | 100% | ORF1ab, ORF8 | 1-Direct sample |
| Novel Coronavirus (2019-nCoV) RT-PCR | DNK-1418-1 | Dynamiker Biotechnology (Tianjin) Co., Ltd. | 10 copies/reaction | 100% | ORF1ab, N, Actin* | 1 |
| PhoenixDx® SARS-CoV-2 IVD | PCCSKU15261 | Procomcure Biotech GmbH | 50 copies/reaction | 100% | RdRp, E | 3 |
| U-TOPTM COVID-19 Detection Kit | SS-9930 | SEASUN BIOMATERIALS | 10 copies/reaction | 100% | ORF1ab, N, RNAse P* | 1 |
| NeoPlex COVID-19 Detection Kit | NR05A | GeneMatrix, Inc. | 50 copies/reaction | 100% | RdRp y N | 1 |
| iAMP® COVID-19 Detection Kit | iAMP-100 | Atila BioSystems | 72 copies/reaction | 100% | ORF1ab, N | 1 |
| genesig® Coronavirus COVID-19 Real-Time PCR Assay | Z-Path-COVID-19-CE | Primerdesign Ltd. | 5 copies/reaction | 100% | RdRp | 1 |
| DiaPlexQ Novel Coronavirus (2019-nCoV) Detection Kit | SQD52-K100 | SolGent Co., Ltd. | 10 copies/reaction | 100% | ORF1ab, N | 1 |
| RIDA ® GENE SARS-CoV-2 RUO | PG6815RUO | R-Biopharm AG | 10 copies/reaction | 100% | E | 1 |
| COVID-19 Real-Time PCR Kit | HBRT-COVID-19 | Chaozhou Hybribio Biochemistry Ltd. | 250 copies/reaction | 100% | ORF1ab, N | 1 |
| Abbott Real Time SARS-CoV | 09N77-090, 09N77-080 | Abbott Molecular Inc. | ** | 100% | RdRp, N | 1- Direct sample |
| CDC 2019-Novel Coronavirus (2019-nCoV) (CDC) | 10006606, 10006625, 10006626, A6121 | Integrated DNA Technologies-Promega Corporation | 5 copies/reaction | 100% | N1 and N2, RNAse P* | 3 |
| AccuPower SARS-CoV-2 Real-Time RT-PCR Kit | SCV-2122 | BIONEER CORPORATION | 10 copies/reaction | 100% | E, RdRp | 2 |
| SARS-CoV-2 Real Time PCR Kit | RTPCR001 | Vircell microbiologists | 10 copies/reaction | 100% | E, N, RNAsa P* | 2 |
| Allplex 2019-nCoV Assay | RP10244Y | Seegene Inc | 10 copies/reaction | 100% | RdRp, N, E | 1 |
| GeneFinder COVID 19 PLUS RealAmp Kit | IFMR-45 | OSANG HEALTHCARE LTD | 10 copies/reaction | 100% | RdRp, N, E, RNAse P* | 1 |
| Logix Smart (COVID-19) | COVID-K-001 | CO-DIAGNOSTICS, INC | 10 copies/reaction | 100% | RdRp, human gene* | 1 |
| Commercial cases LightMix® Modular SarbecoV E-gene EAV, SARS-CoV (COVID19) N-gene, and SARS-CoV-2 (COVID-19) RdRP | 40-0776-96, 53-0775-96, and 53-0777-96, respectively | TIB MOLBIOL, LLC | 10 copies/reaction | 100% | RdRp, N, E | 3 |
| Xpert® Xpress SARS-CoV-2 | XPRSARS-COV2-10 | CEPHEID | ** | 100% | N2, E | 1-Direct sample |
| cobas®SARS-CoV-2 Test | 9175431190 | Roche Molecular Systems | ** | 100% | ORF1, E | 1-Direct sample |
| Detection kit for 2019 Novel Coronavirus (2019- nCoV) RNA (PCR-Fluorescence Probing) | #DA-930 | Daan Gene Co., Ltd. Sun Yat-Sen University | 10 copies/reaction | 100% | ORF1ab, N | 1 |
| VIASURE SARS-CoV-2 | VS-NCO206L | CerTest Biotec, S.L. | 10 copies/reaction | 100% | ORF1ab, N | 1 |
| WoV19 Kit | G2L-WoV19-SP- 100E | GENES2LIFE SAPI DE CV | 250 copies/reaction | 100% | E, RdRp, RNAse P* | 3 |
| DeCoV19 Kit | G2L-DeCoV19-SP | GENES2LIFE SAPI DE CV | 250 copies/reaction | 100% | N1, N2, N4, RNAse P* | 3 |
| DeCoV19 Kit Triplex | G2L-DeCoV19-MP | GENES2LIFE SAPI DE CV | 250 copies/reaction | 100% | N1, N2, N3, RNAse P* | 2 |
| TaqMan 2019-nCoV Assay Kit v1 | A47532 | Applied Life Technologies Corporation, ThermoFisher | 10 copies/reaction | 100% | S, ORF1ab, N, RNAse P* | 3 |
Obtained from the following electronic address (constantly updated) (date of consultation 01-07-2020): https://www.gob.mx/cms/uploads/attachment/file/561223/Listado_de_estuches_comerciales_utiles_para_el_diagn_stico_de_SARS-CoV-2.pdf
*RNAse P and actin genes are human genes that assess the quality of the sample.
**Closed platforms with higher sensitivity than conventional tests.
Pre-analytical phase
The pre-analytical phase is considered the first stage of process development in the clinical laboratory. Deviations or omissions in this stage impact adequate performance, generating failures in the tests performance or even affecting the quality of the results. In the case of SARS-CoV-2 detection, this phase is essential for identifying the virus, diagnosing the disease, and the care of healthcare personnel exposed to such processes.
It is crucial to be aware of biosafety measures during all stages of the analytical process considering all the risks involved in handling biological samples of infectious diseases. The minimum essential personal protective equipment (PPE) is described in the Biosafety and Biosecurity Protocol issued by the InDRE to handle patients during sample collection. Further details can be found in the following link: https://www.gob.mx/salud/documentos/lineamientos-vigentes-red-nacional-de-laboratorios-de-salud-publica?state=published) (Table 2).
Table 2 Minimum essential personal protective equipment to handle patients during sample collection
| Description of the personal protective equipment | Quantity needed for each sample collection |
|---|---|
| Disposable, single-use, waterproof gown with reinforced sleeves and chest, length exceeding the height of the boots | 2 |
| Long-sleeved nitrile gloves or as recommended by the WHO 280 mm (kit or pair) | 2 |
| Hair cover with elastic | 2 |
| Shoe covers (kit or pair) | 2 |
| Surgical gown (optional) | 2 |
| NIOSH N95 or N100 respirator or disposable surgical mask | 2 |
| 3.5 cm wide microporous tape | 1 |
| Safety glasses, preferably with silicone seals | 2 |
| Rigid closed-toe shoe (white rubber sanitary boot when available) (kit or pair) | 2 |
| Supplies for an incident | 1 |
| Product description | 1 |
| Autoclave sterilized paper roll | 1 |
| Red translucent plastic bag for biohazardous and infectious waste, 200 caliber that fulfills NOM-087-SEMARNAT-SSA1-2002 | 1 |
| Gel alcohol 60-80% | 1 |
| Sodium hypochlorite solution 0.05% | 1 |
| Ethanol 70% solution | 1 |
| Triple packaging system Category B | 1 |
| Hermetic container for the transfer of packaged waste to the sterilization facility | 1 |
*Taken from the Biosafety and Biosecurity Protocol issued by the Epidemiological Diagnostic and Reference Institute (InDRE).
NIOSH, National Institute for Occupational Safety and Health; PPE, personal protective equipment; WHO, World Health Organization.
During the development of molecular tests, errors to identify SARS-CoV-2 in the pre-analytical phase are similar to those in other diagnostic areas. An example is the type of specimen recommended for these tests; the primary specimen should come from the respiratory tract of the individual with suspected disease. Nasopharyngeal (NP) and oropharyngeal (OP) swabs should be collected and transported in the same tube containing viral transport medium or sterile saline. Primary samples should be kept refrigerated (4-8 °C) and processed within 24-72 h after sampling.
Both the process and the timing of sample collection are critical to the outcome. By the process, we mean the swabbing technique and the region where it will be performed, while the timing of sample collection refers to the time elapsed after exposure to the virus. The estimated incubation period of this new virus ranges from 2 to 14 days; however, incubation periods of 21, 24, or 27 days have been observed in some cases. Therefore, sampling at the early stages of exposure may generate false negative or indeterminate results in most cases19-25.
Kucirka et al. used a model to estimate the false-negative rate and found that sampling suspected individuals before the onset of symptoms show up to a 100% chance of a false-negative result. This value decreases with the time of symptoms onset. For example, on day 1 of symptoms onset, the estimate reaches 67% (95%CI 27-94%), and on day 4, the mean false negative rate is 38% (95%CI 18-65%). This rate decreased to 20% (95%CI 12-30%) on day 8, and by day 21, the false-negative rate increased again26.
Therefore, the sampling moment should be considered decisive since the incubation cycle of the virus and the manifestations of the disease are variable in each patient and may affect the interpretation of the results (Figure 1).
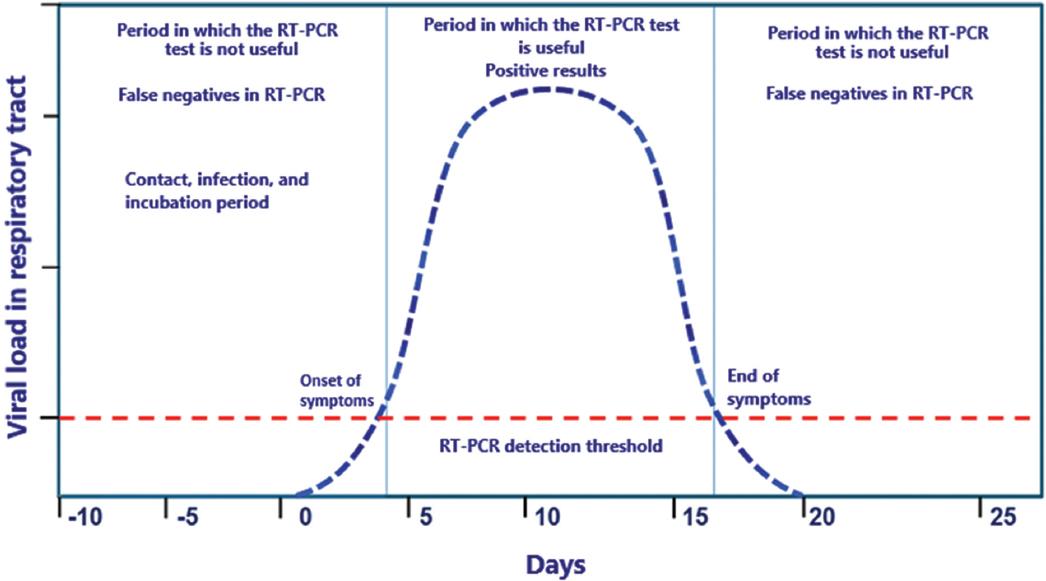
Figure 1 SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) infection clinical course concerning the reverse transcription-polymerase chain reaction (RT-PCR) test positivity.
Another critical variable is the difficulty of sampling in newborns and infants due to the characteristics of the material used for sampling. Although small or low-caliber swabs are available for these patients, the sample collected is usually not representative. Therefore, the personnel in charge of sample collection must be trained in patient-management and sampling material.
For upper respiratory tract sampling, including NP and OP swabs, the following recommendations should be considered:
1) Synthetic fiber (flocked dacron or polyester) swabs, preferably with a flexible wire handle, should be used. As calcium alginate or wood-based swabs may contain substances that affect some viruses viability and inhibit RT-PCR, their use is not recommended. Swabs should be immediately placed in sterile tubes containing viral transport medium (commercial or prepared in the laboratory; e.g., Hanks salts, depending on the diagnostic protocol established or used)10-17. The swab should be cut at the height of the tube to facilitate handling the primary sample during the analytical phase.
2) For NP sampling, it is recommended to insert a synthetic fiber swab with a flexible wire handle through the nares parallel to the palate (not upward) until resistance is encountered or the distance is equivalent to that from the patients ear to the nose, indicating contact with the nasopharynx. The swab should reach a depth equal to the distance from the nostrils to the external ear. Once inside, it is essential to gently rub and rotate the swab for a moment (3 to 5 seconds) to absorb the secretions. Then the removal should be done slowly with a gentle twist. Finally, a visual check should be made to ensure that the swab is moist with the mucosal sample10-17.
3) In OP sampling, the swab should be inserted into the posterior pharyngeal and tonsillar areas, swabbing while the swab is rotated over the tonsillar pillars and posterior oropharynx, avoiding touching the tongue, teeth, and gums. As with NP sampling, it is important to visually verify that the swab is moist with the mucosal sample10-17.
4) It is necessary to be trained in obtaining samples such as sputum, saliva, NP aspirate, or nasal aspirate, and have experience in the handling of vials, containers, and adapters to reduce the risk of contamination, leakage, and generation of aerosols10-17.
5) Personnel must be trained in the fitting and removal of PPE, considering that all material and equipment are disposable (except for safety glasses) and must undergo a sterilization process before disposal.
6) It is recommended to work in pairs, as this facilitates the handling of pediatric patients and manipulating the material.
7) The site for outpatient sampling should be different from the site routinely used for blood and microbiological sampling. Due to the pathogenic characteristics of SARS-CoV-2, an exclusive site for sample collection is essential, considering infrastructure, lighting, ventilation, biosafety equipment, contamination control equipment, and the identification of the bio-hazardous waste-collection route.
8) Samples must be transported with triple packaging specifications. A trackable control of the route, temperature, transport times, and the contacts during the whole process must be recorded.
Analytical phase
For molecular biology tests, it is crucial to recognize two stages within the analytical phase. The first phase involves obtaining the genetic material, which will be the primary source of the process, while the second stage corresponds to the RT-PCR. Before implementing the tests, the following elements are required: the qualification of the facilities and the verification of the chosen analytical platform, in addition to a specific documented operating procedure.
When performing these analytical procedures, the PPE used should be the same as in the pre-analytical phase, and the analytical process should be performed in a biosafety level 2 (BSL-II) laboratory at least10-28.
As previously described, identification of SARS-CoV-2 in the upper and lower respiratory tract is performed by RNA amplification via real-time RT-PCR during the virus infection cycle that allows qualitative detection in vitro. Positive results indicate an active viral infection; however, bacterial infection or co-infections with other viruses are not excluded. Therefore, negative results should be combined with clinical observations, patient history, and epidemiological information to guide treatment and other medical decisions.
We suggest the following recommendations for handling the primary sample and obtaining viral RNA:
1) The sample matrixs handling, previously identified, should be carried out in a BSL-II laminar flow hood; cabinets should not be used in this procedure. All the material with direct contact with the sample should be kept inside the hood and should not be removed until it has been correctly decontaminated with UV light for at least one hour. In addition to the PPE previously described, double pair of gloves is recommended during this procedure.
2) Automated equipment is recommended to obtain viral RNA and reduce the analysts contact with the sample, thus decreasing contagion risk. Also, the concentration and purity of the viral RNA obtained should be registered.
The viral RNA obtained is not infectious; however, it should be kept refrigerated (4-8°C) before the RT-PCR test (which should be performed within 2-3 h after obtaining the RNA) and separate from the reagents used for the test. Once used, this biological material can be stored at -80oC.
For RT-PCR performance, the PPE equipment should be new. It is crucial to consider the following recommendations:
1) The reaction mixtures preparation should be carried out in a different location from where the genetic material and the negative and positive controls will be added. The reaction mixture can be prepared either in a cabinet or in a laminar flow hood.
2) The reagents used in the reaction mixture should be stored separately from the genetic material used as positive and negative controls. The positive and negative controls are synthetic genetic material without infectious capacity and are designed and determined by the choice protocol. In our protocol, we use the ribonuclease p gene (RP gene) as an endogenous gene, while for the detection of SARS-CoV-2, we use the RdRP and E genes (Figure 2).
3) It is preferable to make aliquots of the reagents, considering the number of samples processed routinely, to avoid the continuous freezing and thawing of reagents and samples.
4) Sanitization and decontamination of both work surfaces and equipment (micropipettes, centrifuges, among others) should be performed before and after use. Among the agents that can be used for this purpose are 10% bleach, 70% ethanol, and commercial RNA inhibitors to reduce contamination risk.
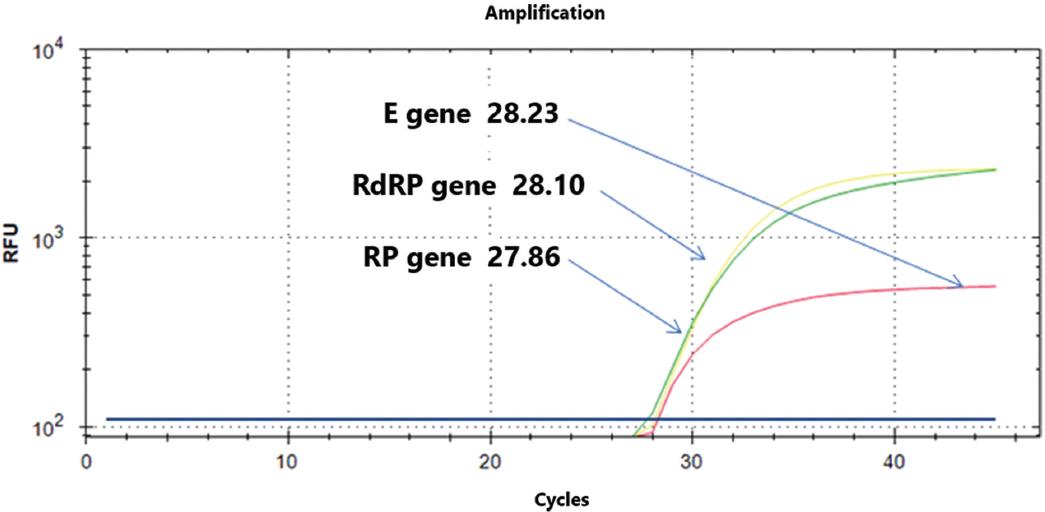
Figure 2 Detection of SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) using the Berlin protocol in a positive case (amplification cycles of the three genes). E gene, envelope protein gene; RdRP gene, RNA-dependent RNA polymerase gene; RP, RNase P gene.
An essential part of the analytical process is diagnostic accuracy. In the tests for SARS-CoV-2 identification, there are different methodological proposals or analytical designs. The most widely used at the international level are the following:
1) The protocol designed by the CDC and the protocol developed by the Institute of Virology of Berlin. Both have been compared in several publications, making evident the complexity of homogenizing the various protocols to have similar analytical precision. For this reason, the minimum operating conditions of each protocol should be evaluated15,16.
2) Xie et al. described that 3% of patients with imaging evidence of COVID-19 (chest CT scan with COVID-19 related damage, ground-glass imaging, or mixed ground-glass imaging with consolidation) initially showed a negative RT-PCR test result for SARS-CoV-2 identification. In this group of patients, test results were positive for COVID-19 after some days in the hospital17.
3) In another series of patients, Ai et al. reported that 88% (888/1014 patients) showed chest CT scans with data of SARS-CoV-2 infection but only 59% (601/1014) with RT-PCR positivity, adding to the patients biological variability18. Despite the assay or test variation, we should clearly understand our platforms analytical performance based on the tools that each laboratory should consider within the internal quality control program.
Post-analytical phase
The recommendations during this phase are the following:
1) Each genes amplification cycles in each sample should be considered concerning the amplification cycle of the positive and negative controls. If possible, generate cut-off points according to the population evaluated.
2) Good transcription of the results to the reporting system and matching patient data (name, age, diagnosis, room, among others) must be verified.
In the case of the graphs or the experiments performed to identify SARS-CoV-2, interpretation criteria should consider each genes characteristics and, if necessary, the presence, absence, or combinations that can be obtained. For example, the Berlin protocol seeks to identify three genes (a constitutive gene, a generic viral gene, and a specific viral gene) (Table 3).
Table 3 Table of interpretation of reverse transcription-polymerase chain reaction (RT-PCR) results
| E gene | RdRp gene (discriminatory) | RNase P | Results |
|---|---|---|---|
| + | + | + | Positive for SARS-CoV-2 |
| + | - | + | Probable Sarbecovirus |
| - | - | + | Negative for Sarbecovirus and SARS-CoV-2 |
| - | - | - | Not adequate assay |
| - | + | + | Repeat/Send to InDRE for advice |
InDRE (Spanish acronym), Epidemiological Diagnostic and Reference Institute; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Currently, no quantitative method can detect the number of virus copies in a sample; however, the number of amplification cycles of a specific gene detected in RT-PCR can provide information on the concentration of viral load (Table 4). It is important to be aware of some considerations to extrapolate these data: a) RNA concentration obtained during the extraction should be homogenized for all tests. Therefore, RNA should be quantified before the RT-PCR test and dilutions should be performed if necessary; b) the gene used for this determination should always be the same; 3) the interpretation will depend on the protocol implemented, the equipment used for the determination, and the brand of reagents selected.
Table 4 Relative estimation of viral load in positive results, using a specific gene (RdRp gene) of SARS-CoV-2 by reverse transcription-polymerase chain reaction
| Amplification cycle | Relative estimate of viral load |
|---|---|
| < 20 | Very high viral load |
| 20-25 | Medium/high viral load |
| 25-30 | Intermediate viral load |
| > 30 | Very low viral load |
SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
In the case of interpretation errors, the final report delivered to the physician should be clear and concrete. In the case of positive results for COVID-19, unnecessary information (e.g., primers sequence, enzymes used, and other methodological details) should not be included in the reports.
Finally, laboratories from public and private institutions that perform SARS-CoV-2 identification tests must generate a report of positive cases to the federal governments epidemiological surveillance network for the follow-up of the pandemic in the country. Each healthcare center must report the results of the SARS-CoV-2 tests to each countrys competent health authorities. In the HIMFG Laboratory, we generate weekly reports following the current strategies, and we are part of the epidemiological surveillance support laboratories.
Molecular detection of SARS-CoV-2 is critical for taking action against the pandemic. However, it is essential to perform these procedures using protocols that promote patients and healthcare personnels safety. In Mexico, the Official Standards (NOM, for its Spanish acronym) for the organization and operation of clinical laboratories (NOM-007-SSA3-2011) and for the management of bio-hazardous waste (NOM-087-ECOL-SSA1-2002), among others, allow the establishment of clinical diagnostic tests.
Considering the heterogeneity of detection protocols, we should promote inter-laboratory networks for the appropriate exchange of information and relevant actions, besides following the guidelines issued by the InDRE to strengthen the timely detection of SARS-CoV-2 in Mexico. Furthermore, laboratories should continue to use the influenza laboratory algorithm recommended by PAHO and WHO for routine surveillance, considering that testing for COVID-19 should be performed only for patients who meet the case definition following epidemiological surveillance strategies.











 nueva página del texto (beta)
nueva página del texto (beta)


