Introduction
Molecular docking is a computational method used to predict the interaction of two molecules generating a binding model. In many drug discovery applications, docking is done between a small molecule and a macromolecule for example, protein-ligand docking. More recently, docking is also applied to predict the binding mode between two macromolecules, for instance protein-protein docking.
Currently, molecular mechanics is the basis for most docking programs. Molecular mechanics involves the description of a polyatomic system using classical physics. Experimental parameters such as charges, torsional and geometrical angles are used to narrow down the difference between experimental data and molecular mechanics predictions (Lopes, Guvench & Mackerell, 2015). Due to shortcomings and limitations of experimental parameters, mathematical equations may often be parametrized on the basis of quantum-mechanical semiempirical and ab initio theoretical calculations. As such, molecular force fields are sets of equations with different parameters with the final purpose of describing the systems. As force fields may use different considerations and simplifications, the description of the system may be inaccurate due to the level of theory involved (classical physics).
Most force fields rely on five terms, all of which have a physical interpretation: potential energy, torsional terms, bond geometry, electrostatic terms, and Lenard-Jones potential (Monticelli & Tieleman, 2013). Examples of prominent force fields are AMBER, GROMOS, MMFF94, CHARMM, and UFF. An in-depth discussion on force fields and their limitations is beyond the scope of this review but the following references provide for further reading (González, 2011; Guvench & MacKerell, 2008; Lopes, Guvench & Mackerell, 2015).
With the use of force fields, molecular and protein modeling was accomplished in the early 1980s. A natural extension of these methods was the modeling of molecular processes such as protein-ligand binding. Two general methodologies were developed. First, the rigid body approach that is closely related to the classic model of Emil Fischer. In this model, the ligand and receptor are regarded as two independent bodies that recognize each other based on shape and volume. The second approach is flexible docking. This approach considers a reciprocal effect of protein-ligand recognition on the conformation of each part (Dastmalchi, 2016). Figure 1 schematizes the two main approaches for molecular docking.

Figure 1: Schematic representation of two main approaches for molecular docking: A) rigid body and B) induced fit.
Over the past 15 plus years, the number of publications associated with molecular docking has increased substantially. Figure 2 illustrates this point. The figure shows the number of publications since the year 2000 indexed in major search engines that are associated with the keyword “docking”. Unfortunately, the increased use in docking has been accompanied with a superficial application to computer-aided drug design (CADD), including the 1-click drug discovery (Saldívar-González, Prieto-Martínez & Medina-Franco, 2017). Therefore, it is important to bear in mind that molecular docking has limitations as any other technique. In other words, it should be used carefully and consciously, complementing its application with other computational and experimental methods.
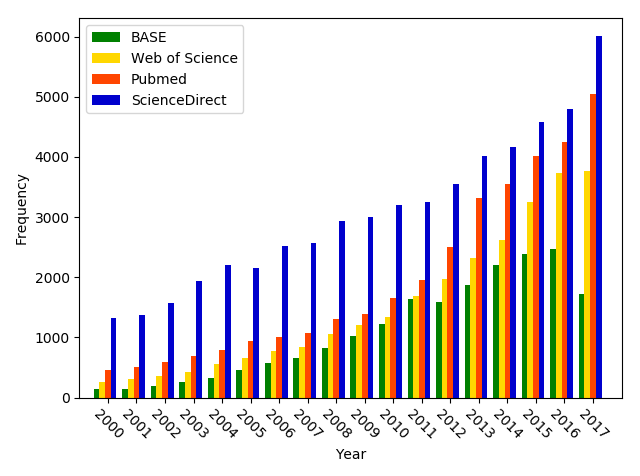
Figure 2: Frequency of papers since 2000 with the keyword “docking” as part of the subject or title. The trends are shown for four major search engines.
There are several servers, suites and programs available for molecular docking. Each tool uses different force fields and algorithms for pose generation, refinement, and calculation of receptor-ligand interactions. Table I presents a short list of major docking programs, including their algorithms and general information.
Table I: Examples of software available for protein-ligand docking and their search algorithm.
| Name | Search algorithm | Type | References |
|---|---|---|---|
| AUTODOCK4 | Lamarckian genetic algorithm | Academic | Morris et al ., 2009 |
| DOCK | Shape matching | Academic | Allen et al ., 2015 |
| OEDOCKING | Shape matching | Academic | Kelley, Brown, Warren & Muchmore, 2015; Mcgann, 2011 |
| FLEKSY | Ensemble-based | Commercial | Nabuurs, Wagener & De Vlieg, 2007; Wagener, De Vlieg & Nabuurs, 2012 |
| SWISSDOCK | Evolutionary optimization | Academic | A. Grosdidier, Zoete & Michielin, 2011 |
| GOLD | Genetic algorithm | Commercial | Jones, Willett, Glen, Leach & Taylor, 1997 |
| GLIDE | Hybrid | Commercial | Friesner et al., 2004 |
| VINA | Local optimization | Academic | Trott & Olson, 2009 |
| RDOCK | Hybrid | Academic | Ruiz-Carmona et al., 2014 |
| LEDOCK | Simulated annealing | Academic | Unzue et al., 2016 |
| PLANTS | Ant colony optimization | Academic | Korb, Stützle & Exner, 2009 |
| HADDOCK | Hybrid | Academic | Dominguez, Boelens & Bonvin, 2003 |
| SURFLEX-DOCK | Shape matching | Commercial | Spitzer & Jain, 2012 |
| MOE | Hybrid | Commercial | Vilar, Cozza & Moro, 2008 |
| FLEXX | Shape matching | Commercial | Kramer, Rarey & Lengauer, 1999 |
| FITTED | Hybrid | Commercial | Corbeil & Moitessier, 2009; De Cesco, Kurian, Dufresne, Mittermaier & Moitessier, 2017; Englebienne & Moitessier, 2009; Moitessier et al., 2016 |
| LIGANDFIT | Shape matching | Commercial | Venkatachalam, Jiang, Oldfield & Waldman, 2003 |
| ICM | Hybrid | Commercial | Neves, Totrov & Abagyan, 2012 |
| IGEMDOCK | Evolutionary algorithm | Academic | Yang & Chen, 2004 |
Despite the fact there are a large number of robust docking programs available, the reader should bear in mind that not all docking algorithms are suited for any given system (Cole, Davis, Jones & Sage, 2017). It is generally advisable to use more than one docking program: different studies have shown that, overall, taking a consensus from various docking protocols yields better assessment of protein ligand interactions and more reliable pose ranking (Houston & Walkinshaw, 2013; Tuccinardi, Poli, Romboli, Giordano & Martinelli, 2014).
The goal of this review is to discuss crucial aspects of molecular docking. Challenges and perspectives are also commented.
General recommendations and guidelines for docking
Hardware and software requirements for molecular docking
Before addressing the scientific details of the docking methodology, we will comment on the general hardware requirements to run docking efficiently. Usually, a docking calculation is not considered CPU intensive as ligands may be docked and evaluated in a couple of minutes. Currently, almost any personal computer (or laptop for that matter) is competent enough to run a small docking campaign (around 500-1000 compounds) in a reasonable time. However, docking-based virtual screening of public repositories may escalate quickly (more than 106 compounds), requiring more computing resources to finish in a couple of weeks. Table II presents general guidelines recommended for a computer before running docking.
Table II: Hardware recommendations to run molecular docking simulations.
| Specification | Minimum | Recommended |
|---|---|---|
| Processor architecture | i686 | x86_64 |
| Processor clock rate | 1.5 GHz | 3.4 GHz or higher |
| Number of threads | 2 | 8 |
| RAM | 1 GB | 4GB or higher |
| HDD | 1 GB | 10 GB or higher |
In general, GPU computing has become more efficient and attractive for intensive calculations yielding results at a 5-10-fold increase when compared to CPU-based calculations. Such approaches are well-known in computer-aided design (CAD).
The most notable example and success case for this are molecular dynamics, as many pioneering efforts were made to make these calculations scalable. Noteworthy examples include AMBER, GROMACS, Desmond, NAMD and CHARMM, all of which, have been ported to make use of Compute Unified Device Architecture (CUDA) developed by NVIDIA Corporation. With the use of GPUs, a workstation may process the same amount of information as a CPU-cluster, enabling the simulation of large systems or conducting longer simulations. Following the success of these approaches, other methods were optimized for CUDA: ab initio (GAMESS, Firefly), semi-empirical calculations (MOPAC), FEP calculations (Desmond), and similarity searching (FastROCS).
In this regard, what is the status of molecular docking? After all, molecular dynamics is a distant relative and is GPU optimized. Most molecular dynamics running on GPUs use a split method, which leaves the calculation of non-bonding interactions to the GPU while the CPU deals with constraints and electrostatics (Krieger & Vriend, 2015). Unfortunately, due to methodological differences molecular docking has not seen a true GPU implementation. Searching algorithms and scoring schemes involved in docking may be, in principle, implemented in GPUs. However, the programming and breakdown of such algorithms is not straightforward (Kannan & Ganji, 2010). Additionally, some docking software do not consider GPU optimization entirely such as Vina.
While it is true that an academic researcher rarely will need to dock more than 105 compounds, virtual screening would improve with faster technologies. A clearer application can be seen in reverse virtual screening or target fishing. This method involves docking of a set of one or few ligands against a broad array of protein families, with the focus of identify a potential target or polypharmacology profile (Cereto-Massagué et al., 2015; Lavecchia & Cerchia, 2015; Medina-Franco, Giulianotti, Welmaker & Houghten, 2013; Méndez-Lucio, Naveja, Vite-Caritino, Prieto-Martínez & Medina-Franco, 2016).
GPU-optimized docking may become a standard in the near future. Nonetheless, at present time such attempts are more exception than a rule. Examples include DOCK6 with GPU implementation, which allows for faster calculation of its AMBER scoring function (Yang et al., 2010); Molegro Virtual Docker which also used CUDA for GPU support and beta builds for Autodock 4 (Altuntaş, Bozkus & Fraguela, 2016; Mendonça et al., 2017).
Is there such thing as the best molecular docking program?
Common questions in the field are: what is the best docking program? With plenty of choices and approaches (e.g., Table I), were to start? Academic or free software is a good starting point, due to ease of access. Unfortunately, most of these programs have a steep learning curve (e.g. DOCK, rDock and PLANTS). One of the reasons is the lack of penetration of Linux and UNIX-like operating systems in the computer literacy of average users. That is not to say that docking software is exclusive to UNIX-based OSs, just that some programs will depend on interpreters like Cygwin or Git to run properly. Nonetheless, the installation and use of such tools may prove troublesome, especially to novice users. In other cases, docking programs are not designed to run on Windows. Hence, several “beginner troubles” could be avoided using Linux from the start.
Nonetheless, if the reader wants a Windows-friendly approach, Autodock and Vina (along with AutodockTools) are available for this platform. Furthermore, a very useful GUI may be found on PyRx (Dallakyan & Olson, 2015), with the added benefit of virtual screening capabilities out of the box. Another option for Windows is LeDock which also comes with a simple yet effective GUI (although virtual screening has been implemented for Linux).
Regarding commercial software, the authors have some experience with the programs: MOE, Glide and ICM. Of these, in our experience we consider MOE as one of the most userfriendly with a robust set of tools beyond docking.
Concerning the performance of each program, several studies have been conducted trying to identify “top-tier” programs. Of note, most docking programs while accurate, only achieve an overall success of 65-70%. Recently, a notable comparison of 10 docking programs was made (Wang et al., 2016), comparing the pros and cons of the software evaluated against different benchmarks. This reference serves as a general guideline to choose a docking program for a particular use.
We have commented that there is a higher success rate if more than one program is used. However, as discussed below, the selection for consensus among different programs must be carefully made to avoid false positives. Moreover, it is important to remember that docking software was also validated against a training set. As such, some systems may be outside of its applicability domain. Based on these points, the authors make the following albeit subjective recommendations for different docking software:
Vina is an excellent starting point and may be seen as a “jack of all trades” due to a good performance against several protein families. It has various derivatives and forks which provide additional features and ease of usage.
LeDock is a relative new program. Its simple use and quick calculations are its major strengths. Also, poses show a significant clustering and it comes with LePro that is a preparation module. Its weaknesses are the binding energy values and the inability to change the number of docking poses.
MOE has a nice GUI and it is very intuitive. For docking it has several search algorithms and scoring functions. Furthermore, MOE offers support for additional tools such as MOPAC, GAMESS, Gaussian, NAMD, FlexX, GOLD and OMEGA. Its weaknesses are unexpected results with its protein preparation module as it yields wrong charges in some cases.
DOCK is one of the first docking programs developed. Its current version DOCK6, introduces new scoring functions and analysis methods. As mentioned before this is one of the few programs with GPU implementation for AMBER scoring and PBSA/GBSA calculation for protein-ligand complexes. As it is designed to work with DockPrep, the download and usage of USCF Chimera is required, nonetheless this allows the introduction of yet another very useful and efficient tool. The relative downsides of DOCK is that it is only implemented in Linux, it requires some experience in software compilation and relative underperformance of its default scoring function when compared to other programs.
rDock is robust Linux-based program with significant performance. Its steep learning curve may be one of its major weaknesses, as well as it may give troubles if the end-user has never compiled from source. Additionally, rDock does not have a preparation module.
PLANTS has a well balance between usage and performance. It allows for water displacement calculations. The downside are its lack of GUI and steep learning curve. A good alternative is to use Vega ZZ (Pedretti, Villa & Vistoli, 2004) in Windows as its GUI, for easier use. This GUI also provides support for rescoring poses from other docking programs using PLANTS.
Theory of molecular docking
The docking process may be divided on three main parts: 1) preparing the ligand and macromolecule. This is made based on force fields allowing for surface representation and cavities as potential ligand sites (Halperin, Ma, Wolfson & Nussinov, 2002); 2) defining the docking type: rigid or flexible (Agarwal & Mehrotra, 2016); and 3) setting the search strategy for ligand conformations: systematic or stochastic (Ferreira, Dos Santos, Oliva & Andricopulo, 2015). Each part is further elaborated on next sections.
Ligand and protein preparation
The system must be carefully selected and prepared before doing any calculations. The first step is to obtain a structure of the protein, preferably with a bound ligand. Which structure should be used? It is highly recommended to consider three-dimensional structures with high resolution or structures co-crystallized with high affinity ligands or natural substrates. For some proteins, this may not be always the case. In such instances, structures with previous reports of docking or structural studies may be used.
As mentioned in the introduction, docking requires the assignment of several parameters. The information contained on a file of the Protein Data Bank (PDB) is often insufficient and therefore the rectification of PDB files is required. In practice, several preparation modules are available (see Table III), most of them can correct common problems in PDB files. However, it has been shown that some structural aspects are often overlooked (Warren, Do, Kelley, Nicholls & Warren, 2012).
Table III Software used for protein and ligand preparation.†
| Software | Type | Features |
|---|---|---|
| MOE | Commercial | Correction of residue issues, structure clean-up, charge assignment based of several forcefields, protein minimization and binding site prediction. |
| Maestro | Academic/Commercial | Correction of residue issues, structure clean-up, charge assignment, tautomer assignment, loop remodeling*, binding site prediction.* |
| YASARA | Academic/Commercial | Correction of residues, binding site analysis, contact analysis, loop remodeling*, charge assignment*, protein minimization* and hydrogen bonds optimization.* |
| Lead Finder | Commercial | Structure optimization, charge assignment, rotamer selection |
| RosettaLigand | Academic | Structure optimization, charge assignment, rotamer selection, loop optimization. |
| BALLView | Academic | Protein minimization and charge assignment. |
| DeepView | Academic | Protein optimization, loop remodeling and binding site analysis. |
| Vega ZZ | Academic | Correction of residue issues, structure clean-up, charge assignment (forcefield/gasteiger), semiempirical charges and protein minimization. |
| SPORES | Academic | Structure preparation, geometry optimization, connectivity correction and tautomer assignment. |
| UCSF Chimera | Academic | Structure clean-up, charge assignment, loop remodeling, protein minimization. |
| Autodock Tools | Academic | Structure clean-up, charge assignment (Gasteiger), rotamer selection and binding site prediction. |
| Openbabel | Open source | Charge assignment, multiple file formats supported, file conversion. |
† Presented as illustrative software, due to well-known capabilities or ease of use; for other applications of general use please refer to Prieto-Martínez & Medina-Franco, 2018*These features require commercial licensing.
The selection of the parametrization method depends on the software; for example, Autodock and SwissDock use an in-house force field, whereas MOE and LeDock use AMBER and CHARMM charges and atom types, respectively. Of note, the modules of protein preparation of different programs create slightly different output files having a direct impact on the docking results (Feher & Williams, 2012). Consequently, to compare results of docking (e.g., in virtual screening) it is strongly recommended to use the same preparation protocol for all the docking calculations.
Usually, ligand and protein may be processed and prepared separately. Ligand preparation involves similar considerations, while the first step often involves its extraction from the protein structure. In virtual screening, ligands may come from sources different from PDB (e.g. Public repositories like PubChem, organic synthesis or virtual compounds). In such cases, the procedure may vary, but it involves the construction of such molecule from its Simplified Molecular Input Line Entry (SMILES) format or sketching the molecule and save it in a SDF/MOL file, which serves as input. Alternately, following molecule construction several optimizations may follow: geometry and/or charge assignment using ab initio or semiempirical methods, energy minimization or conformational/tautomer search.
It is worth mentioning that the protein may also be prepared on a similar manner (using semi-empirical calculations (Stewart, 2008; Wada & Sakurai, 2005). At first this may seem impractical due to considerable computing times. However, it has been shown that complex optimization and the use of detailed parameters significantly improve results and modelling of interactions (Hostaš, Řezáč & Hobza, 2013; Nikitina, Sulimov, Zayets & Zaitseva, 2004; Ohno, Kamiya, Asakawa, Inoue & Sakurai, 2001).
Finally, we encourage the visual inspection of the output of protein and ligand. This is because the preparation method can sometimes lead to errors in molecular description i.e., wrong connectivity, missing bonds, abnormal geometries, etc. Such errors tend to happen more often when converting between molecular formats and therefore are easily propagated (Feher & Williams, 2012). One most “use with caution” steps are modules implemented in some software packages that enable the user to prepare the structure of the ligand with one or few clicks. These modules should not be used as black boxes. Madhavi et al. showed that careful selection and refinement of the structures for docking led to improve the results of virtual screening (Madhavi Sastry, Adzhigirey, Day, Annabhimoju & Sherman, 2013).
Following the ligand and protein preparation, the binding site must be selected and delimited. This step can be done manually by specifying the coordinates, or automatically using the coordinates of any bound ligand. Additionally, some programs allow for the calculation of cavities or probable binding sites. This is especially useful in cases when the binding site is not known. Also, some programs are capable of blind docking i.e., the search space involves the entirety of the macromolecule. Such approach has been used for the identification of putative binding sites (Marzaro et al., 2013) or allosteric sites (Espinoza-Fonseca & Trujillo-Ferrara, 2005).
The mapping of the binding site (where the docking calculations will be centered) is often done by the GRID methodology (Goodford, 1985). Briefly, the grid can be considered as a box of given dimensions divided on smaller cubes in which a probe atom delineates a possible interaction contour. Therefore, the results are dependent of the GRID resolution and size. Recently, Feinstein & Brylinski (2015) studied the influence of box size on the identification of hits and virtual screening time, as this may be done on the fly or recalculated for each ligand. A good example to illustrate this point are Autodock and Vina: the former needs the precalculation of GRID for molecular docking while the latter does it on the fly yielding a faster calculation (Trott & Olson, 2009).
Defining the type of docking
Following protein and ligand preparation, the type of docking must be selected. In the case of flexible docking, residues are then selected as additional terms for energy calculation. This may be done manually (Autodock, Vina) or automatically (ICM, MOE). This usually means the potential is “softened” to allow residue flexibility and its interaction with the ligand (Dastmalchi, 2016). Briefly, softening the interaction potential involves any modification to the parameters of interaction description. Frequently this involves the Lennard-Jones potential (Equation 1), as it description for steric clashes may be too conservative and cannot sample subtle changes that can happen in a protein pocket. Therefore, if such term is reduced (e.g. 9-6 instead of 12-6) it can account for protein flexibility allowing more ligand conformations (Ferrari, Wei, Costantino & Shoichet, 2004).
Lennard-Jones potential, as implemented in GRID-based simulations
Setting the search strategy for ligand conformations
The ligand conformation and placement will depend on the selection of the docking algorithm. This involves a conformational search and the selection of the optimal solution per the scoring function. Such search may be systematic or at random.
Systematic search involves a comprehensive sampling of conformations and combination of structural parameters. This approach, however, makes use of more resources and takes significantly more time to construct the conformers and evaluate them individually. Additionally, a true systematic search may generate a combinatorial explosion. To avoid this problem, systematic search is done by building the ligand from different fragments: selecting one as anchor and sequentially adding combinations of remaining fragments (Ferreira et al., 2015).
Stochastic search is made randomly by means of two main methods: Monte Carlo (MC) or genetic algorithms (GA). Each develops different conformations based on bond rotations as degrees of freedom (Meng, Zhang, Mezei & Cui, 2011). Structures are then submitted to the scoring function for pose selection and filtering.
Scoring functions
The conformational search discussed above may yield a large number of structures. Of course, only a small percentage of these may be biologically relevant. Scoring functions are then used to distinguish good from bad solutions evaluating a broad range of properties including, but not limited to, intermolecular interactions, desolvation, electrostatic, and entropic effects (Ferreira et al., 2015). Scoring functions can be classified as force field-based, empirical, and knowledge-based.
Force field-based
This type of scoring function uses the parameter contribution to construct a “master function” (e.g., Equation 2), which account for bond stretching, electrostatics, non-bonding interactions, etc. In turn, this has the limitation that entropic contributions of solvation cannot be accounted for (Kitchen, Decornez, Furr & Bajorath, 2004). Moreover, these approaches usually involve longer computing times and need of distance cutoffs decreasing the accuracy of long-range effects (Meng et al., 2011).
Example of a hypothetical force field-based scoring function
Empirical
Empirical scorings functions involve the reproduction of experimental values. In this regard they may be related to QSAR models: linear regressions of descriptors (Eldridge, Murray, Auton, Paolini & Mee, 1997) to model protein-ligand interactions (Pason & Sotriffer, 2016). Therefore, the function is simpler. See for example the PLANTS score in Equation 3. Briefly, PLANTS scoring involves the evaluation of shape complementarity based on atom types as classified from training sets (f PLP), ligand clash potential (f clash ), ligand torsion potential as implemented by Tripos forcefield (f tors ), and site contributions to enforce ligand interactions (c site ). The value -20 is introduced for score shifting (Korb et al., 2009). However, it has been argued that such energy parameters may not be robust enough (as their QSAR counterparts (Ferreira et al., 2015; Meng et al., 2011). Furthermore, most empirical scorings suffer significant limitations, as they are derived from individual protein-ligand complexes and heterogeneous data in training sets (Pason & Sotriffer, 2016). Still, empirical scoring functions are computed much faster than their force field-based homologues and yield reasonable binding energy predictions (Murray, Auton & Eldridge, 1998).
Empirical scoring function as implemented by PLANTS
Knowledge-based
These scoring functions are designed to reproduce structures and not energies. The structure is constructed using pairwise potentials derived from known receptor-ligand complexes (Ferreira et al., 2015; Kitchen et al., 2004). This is based on the inverse Boltzmann relation (vide infra), which comes from statistical mechanics and as such involves mean force potentials instead of real ones (Huang, Grinter & Zou, 2010). For example, consider the Equation 4. It shows the score calculated by Small Molecule Growth (SMoG) 2001. The first term involves the normalization of contact probability between protein and ligand, F rotX refer to the contribution of “frozen” flexible bonds, which reduce conformational entropy of ligands, and N rotX denotes the number of flexible bonds of types I and II (Ishchenko & Shakhnovich, 2002).
Knowledge based scoring function as implemented by OpenGrowth
A significant advantage of knowledge-based functions is their balance between performance and calculation time. Additionally, these can consider uncommon interactions e.g., sulphur-aromatic (Meng et al., 2011). On the other hand, a limitation of these scorings functions is their reliance on the inverse Boltzmann relationship. A reference state needs to be defined i.e., a state in which pairwise potentials are zero. Defining such state is not trivial and it can impact the results significantly (Muegge, 2000). Attempts to improve the predictive power have resulted in “hybrid” approaches, e.g. SMoG2016 combines empirical data and knowledge-based potentials (Debroise, Shakhnovich & Chéron, 2017).
Docking accuracy
The scope and capabilities of docking have been debated over the years. To date, several benchmark studies comparing different docking programs have been published (Cross et al., 2009; Elokely & Doerksen, 2013; Perola, Walters & Charifson, 2004; Tuccinardi et al., 2014; Wang et al., 2016). It has been shown that, on average, docking methods have around 60-75% success rate on the identification of correct poses (Cole et al., 2017; Wang et al., 2016). A major challenge continues to be the accurate prediction of the energy interaction between two molecules. Thus far, quantitative correlations between experimental activities and docking scores are, in general, low, due to the high level of approximations implemented in scoring functions (see previous section). However, qualitative correlations are quite acceptable as proven by the success of several docking-based virtual screening campaigns.
It often happens that the pose with the top ranked score is not necessarily the best pose (Waszkowycz, Clark & Gancia, 2011). Consider the following example: bromodomains are small proteins with conserved motifs, which are responsible of gene expression/suppression. A very potent inhibitor of BET bromodomains is IBET762, a triazolazepine ligand which has an IC50 in the nanomolar range. Using Vina for redocking using PDB ID 3P5O, the “best pose” is in a completely different orientation compared to the crystallographic reference (Figure 3).
Another issue is the performance in cross-docking: docking of reference ligands to non-native structures of the same protein often results in the wrong prediction of the binding mode (Corbeil & Moitessier, 2009). Different strategies have been pursued to increase the docking accuracy. Two major approaches are ensemble and consensus docking that are discussed in the next two sub-sections.
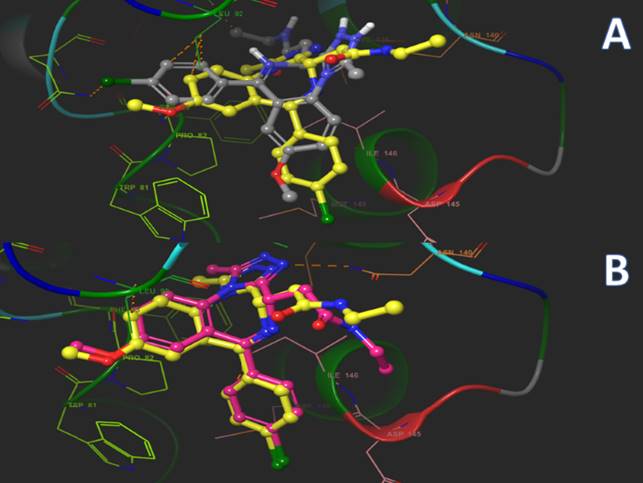
Figure 3: Illustration of docking accuracy. Binding poses obtained from a docking run using Vina with a BET bromodomain (BRD4) and IBET762 (carbon atoms in yellow). A) Best result obtained from scoring (carbon atoms in grey). B) Correct pose for validation (carbon atoms in pink).
Ensemble docking
One of the early applications of ensemble docking was published by Knegtel, Kuntz & Oshiro (1997). Ensemble docking is done by performing multiple docking simulations on different protein conformations. This approach partially accounts for protein flexibility. The main premise is that if a given ligand scores well on a native structure it should do so against a group of conformations allowing the better elucidation of conserved interactions. As such, ensemble docking can be considered as four-dimensional (4D) docking and success rates of 78% of correct binding geometry have been reported (Bottegoni, Kufareva, Totrov & Abagyan, 2009).
Ensembles of conformations of the target molecule may come from crystallographic data, NMR, or computational modeling e.g., molecular dynamics or different homology models. The effects that may influence the performance of ensemble docking are scoring functions (Elokely & Doerksen, 2013), the construction of the ensemble receptors (Craig, Essex & Spiegel, 2010), ligand flexibility (Huang & Zou, 2007), molecular similarity, and enrichment (Ellingson, Miao, Baudry, & Smith, 2015; Korb et al., 2012).
Ensemble docking also serves as a gateway technique for more complex simulations, for instance, solvation. Using three-dimensional (3D) data is possible to make a more in depth approximation of induced fit, which may be integrated with other methodologies like QSAR (Lill, 2007). Co-solvation has proven significant during ensemble construction, as it has shown improved results in virtual screening. Still, it is not clear if such effect is due to the nature of the chemical probes used as co-solvent during model selection (Uehara & Tanaka, 2017).
Ensemble docking has proven successful in a broad range of applications such as identification of hot-spots for protein-protein interaction (Grosdidier & Fernández-Recio, 2008), GPCR modeling (Vilar & Costanzi, 2013), modeling of apo and holo structures (Motta & Bonati, 2017), hit identification (Kapoor et al., 2016), characterization of nuclear receptor modulators (Park, Kufareva & Abagyan, 2010), phase I metabolism (Harris et al., 2014), and toxicity (Evangelista et al., 2016).
Consensus docking
As commented in the introduction, it is recommended using more than one validated program for virtual screening. Given the different scopes of scoring functions and algorithms it is expected that they complement each other. However, since scoring functions are constructed with different bases their comparison and combination is not straightforward (Huang, Grinter & Zou, 2010). A first attempt was made by Charifson, Corkery, Murcko & Walters, 1999. In that study, authors showed a consistent improvement using three independent scoring functions (CHEMSCORE, PLP, and DOCK). Follow up publications suggested that combining three to four independent scoring functions are enough to improve results (Wang & Wang, 2001). An alternative approach was to develop new scoring functions that combined existing methods: premier examples are DrugScore and X-Score (Wang, Lu & Wang, 2003). While this strategy proved useful on certain cases it still has predictive power below average towards highly flexible ligands (Kitchen et al., 2004).
Further efforts in consensus docking focused on improving binding forces and pose selection (Plewczynski, Łażniewski, Grotthuss, Rychlewski & Ginalski, 2011). Certainly, consensus scoring improves pose selection but directly depends on the scores that are combined e.g., a strong correlation among them may increase error rate (Meng, Zhang, Mezei & Cui, 2011). In addition, scoring functions are sensible to specific features of the binding sites (Chaput & Mouawad, 2017).
Alternate approaches towards consensus docking are rank-based and intersection (Feher, 2006) that use statistical criteria to assess the contribution and significance of binding poses. These methods allow pose enrichment and better ranking results (Du, Bleylevens, Bitorina, Wichapong & Nicolaes, 2014; Kukol, 2011) and have the advantage that do not require other input or calculation beyond statistics. Successful applications include hit identification towards HIV reverse-transcriptase (Samanta & Das, 2017), tubulin inhibitors (Fani, Sattarinezhad & Bordbar, 2017), and Zika virus (Onawole, Sulaiman, Adegoke & Kolapo, 2017).
Knowledge-based consensus is a more rational and heuristic approach than rank-based. This strategy requires previous information as it is based on binding mode and significant interactions. Protein-ligand interaction fingerprints (PLIFs) are used to analyze docking results from different programs, searching meaningful interactions and visually inspecting poses to account for clustering or conserved conformations. Figure 4 shows a consensus PLIF for a flavonoid ligand, using four different docking programs: ICM, LeDock, MOE and PLANTS. Again, the bromodoain BRD4 is taken as an example in this figure. For BRD4, three hotspots are known: WPF shelf (W81, P82 and F83), ZA channel (residues 85 through 96) and the AC binding module (Y96 and N140). The histogram presents the frequency of interactions with each amino acid residue as obtained with all four docking programs. The hypothesis is that the example ligand in Figure 4 having consensus interactions is more likely to bind to the WPF shelf and ZA channel. Upon close inspection, the example molecule is similar to flavopiridol, a kinase inhibitor which also binds to BRD4. Due to his binding, flavopiridol is known as a ZA inhibitor and therefore it can be stated that flavonoids conserve such arrangement (Prieto-Martínez & Medina-Franco, 2017; Dhananjayan, 2015).

Figure 4: Knowledge-based consensus using protein ligand interaction fingerprints (PLIFs) and the results of three independent docking runs using different software. A) Results from LeDock. B) Results from ICM. C) Results from PLANTS. D) Results from MOE. E) Consensus interactions.
Although this method is not well-suited for virtual screening, it is useful for filtering and selecting hits (Medina-Franco, Méndez-Lucio & Martinez-Mayorga, 2014; Ran et al., 2015).
Validation
As any technique, docking protocols need to be validated (Verdonk, Taylor, Chessari & Murray, 2007). This process begins before any simulation by protein modeling and selection. As mentioned above, docking input may come from different sources, however the most common is PDB (Berman et al., 2000) that contains over 100,000 structures. Nonetheless, structures in PDB may have some issues such as redundancy, missing atoms, missing residues, and ambiguity.
After preparing the tridimensional structure of the molecular target vide supra, a common practice for validation of the docking protocol is redocking the reference ligand. This is made to test if the docking algorithm produces a correct pose and if the scoring function can identify it as a top pose. As a standard or common measure of validation researchers use the root mean squared deviation (RMSD) (Equation 5) which compares the coordinates of the predicted vs. initial conformation of all heavy atoms of both conformations (Dias & de Azevedo Jr., 2008). Overall, RMSD values should be less than 2.0 Å. A major deviation indicates that the docking program is not suited for prediction of the docking pose.
Root mean squared deviation for two sets of N atoms on c and d coordinates
The implementation of the RMSD calculation is simple and gives direct comparison of reference and prediction. However, this metric is heuristic (Waszkowycz, Clark & Gancia, 2011) and should not be considered as absolute. A more systematic approach is to repeat a redocking at least 50 times, construct a pairwise comparison of reference vs. pose and cluster poses based on a threshold (Morris & Lim-Wilby, 2008). This is done to verify docking convergence. In addition to RMSD, correct pose assessment should be based on interaction-recovery, meaning protein-ligand interactions should match for reference and prediction.
Another quantitative assessment of the performance of docking protocols, in particular for virtual screening, is the analysis of the Receiver-Operating Characteristic (ROC) curve. ROC analysis was developed initially for military applications but it has been adapted to evaluate the performance of screening and diagnostic tests (Zou, O’Malley & Mauri, 2007). ROC curves (Figure 5) plot sensitivity against 1-specificity. An ideal curve must pass over the top left corner, whereas a diagonal over the axes represent random chance. Therefore, the ROC curve allows evaluation of discrimination power independent of event rates (Brown & Davis, 2006). In molecular docking, this validation requires an equal proportion of binders and non-binders. A threshold is then established for scoring, using binders for true positive rates (sensitivity) and non-binders for false positives (1-specificity) (Sørensen, Demir, Swift, Feher & Amaro, 2014). The area under curve (AUC) will represent the probability of ranking random active compounds higher than random inactive ones (Cross et al., 2009; Englebienne & Moitessier, 2009; Park, Kufareva & Abagyan, 2010).
Current challenges
The following sections address crucial topics in molecular docking and some perspectives on the field.
Protein-ligand flexibility and binding evaluation
This is one of the main challenges in docking. Arguably, a proper approach to test the behavior of a protein-ligand complex is in a dynamic setting. Since the introduction Emil Fischer’s model for binding, with more efficient tools and methods it has become clear that subtlety is the protein way. After all, ligand promiscuity has been one of the major questions in medicinal chemistry and pharmacology. With the exponential growth of repositories like the Protein Data Bank (PDB; Berman et al., 2000), new folding patterns and structural arrangements were discovered. This information allows the search for patterns and similarities in binding sites and protein pockets to shed some light into the inner workings of proteins (Ehrt, Brinkjost & Koch, 2016). The “pocket dynamics” has been classified in five classes, which could coexist in a single protein at the same time. This would allow bivalent binding which may serve for selectivity (Stank, Kokh, Fuller & Wade, 2016).
To address the phenomena of induced fit in automated docking we have discussed the use of potential softening to sample side-chain flexibility. In the next sections we pitch some alternatives to advance this problem.
Protein-ligand docking
Structure-based design can be considered an “old-school” yet very powerful approach to druggable targets. In computational methods this is where docking proves the most useful to input many ligand modifications and provide results on the fly. Induced fit docking, while useful, offers a narrow sampling space, as only neighboring residues are considered as flexible and some pocket dynamics or even allosteric effects cannot be considered.
One method to sample flexibility is Protein Energy Landscape Exploration (PELE). This is a Monte Carlo approach which combines protein and ligand perturbations. Basically, three main steps are distinguished: 1) ligand and protein perturbation (using a rotamer library); 2) side-chain sampling (via algorithms) and 3) minimization and acceptance using the Metropolis criteria. Of course, this can prove more expensive computationally but acts as a useful refinement, in particular if the binding site of a protein is not known. Recently, a study showed the successful application of PELE for the correct assessment of binding sites and poses for very flexible proteins (Grebner et al., 2016).
Another take on the problem is by means of machine learning. Using an ensemble of snapshots from molecular dynamics it is possible to apply techniques like self-organizing maps (SOMs) or k-means to determine the complementarity of protein and ligand conformations (Sheng et al., 2014; Vahl Quevedo, De Paris, Ruiz & Norberto De Souza, 2014).
Ensemble docking has prevailed as a notable methodology to sample flexibility. Similar to PELE, Monte Carlo methods have been applied to generate reliable ensembles. One example is MTflex, which uses Monte Carlo Integration to calculate free energy. Building rotamers for binding residues based on low-energy values along the free energy surface, the difference towards other techniques (i.e., rotamer libraries) is that the atoms are added based on a local energy criterion and not to match a structural query. Moreover, reference atom positions may be used to improve results. With this method more efficient ensembles were generated and diminished RMSD notably (Bansal, Zheng & Merz, 2016).
Metadynamics in other strategy to sample protein-ligand flexibility. Metadynamics belongs to the enhanced-sampling techniques in MD simulations. These methods were developed as a workaround the scalability of MD. Metadynamics use biased potentials applied to a selected number of variables in the system (also known as collective variables) to explore deeper into the configurational space of the system (Barducci, Bonomi & Parrinello, 2011). Hence, metadynamics explores conformations outside of local minima using restraints to construct a sum of Gaussian potentials. These potentials are repulsive in nature and have been used before in docking by algorithms such as taboo search. Nonetheless the difference is that metadynamics allows the reconstruction of the free energy surface of the whole system (Gervasio, Laio & Parrinello, 2005). Therefore, metadynamics may be used to validate a binding mode in a more rigorous manner. For example, an induced-fit/metadynamics protocol showed an improvement in Glide performance to identify true positives (Clark et al., 2016).
Docking peptides or peptide-like ligands
Due to their high flexibility and free rotation, peptide sampling is highly variable and virtually irreproducible. Currently, peptides are in medicinal resurgence with examples such as romidepsin (Klausmeyer, Shipley, Zuck & McCloud, 2011; VanderMolen, McCulloch, Pearce & Oberlies, 2011), tacrolimus (Dheer, Jyoti, Gupta & Shankar, 2018) and dolastatin (Senter & Sievers, 2012). Proving that although promiscuous, they can have interesting polypharmacological effects (Díaz-Eufracio, Palomino-Hernández, Houghten & Medina-Franco, 2018) and are attractive as modulators of protein-protein interactions (Díaz-Eufracio, Naveja & Medina-Franco, 2018). Due to the high flexibility, peptide docking involves more exhaustive calculations. To circumvent this impediments several attempt have been made: 1) the use of templates extracted from monomeric ensembles and filtering by energy restraints (Alogheli, Olanders, Schaal, Brandt & Karlén, 2017; Dominguez et al., 2003; Yan, Xu & Zou, 2016), 2) applying restraints and optimizing orientation using local search (Sacquin-Mora & Prévost, 2015), 3) simulated-annealing molecular dynamics and anchor-spot detection on the binding site (Ben-Shimon & Niv, 2015) and 4) systematic selection of rotatable bonds based on latin squares approach to identify energy minima (Paul & Gautham, 2017).
Protein-protein docking
One of the first attempts to model protein-protein docking was Critical Assessment of Protein Interactions (CAPRI; http://capri.ebi.ac.uk) as a contest-space to challenge different human groups, software and servers into correctly predict the conformation of interacting protein-protein pre-chosen targets. The experimental structure of the complex is not know to contestants, but it is to the judges. From the various rounds of CAPRI interesting observations have followed. Indeed, the interaction phenomena between proteins is quite complex even for small changes (Bonvin, 2006). Conceptually, protein-protein docking can be approached as a prediction for the whole complex minimizing each protein by coarse grain models and using local search for the binding sites. However, the local minima of such event involves a multidimensional calculation, whereas the algorithms treat the problem as a global optimization that may not be physically accurate (Vakser, 2014). Thus, the major challenge for protein-protein docking is the flexibility of the backbone. To this end, Monte Carlo techniques have been applied successfully by Rossetta (Wang, Bradley & Baker, 2007). Nevertheless, the bound state of proteins still poses an unsolved problem due to wrong sampling (Kuroda & Gray, 2016). For this reason, comprehensive computational studies need to be conducted to successfully distinguish realistic complexes from unrealistic predictions (Vreven et al., 2015).
Pose vs scoring
Hit selection is an ongoing challenge in docking. As discussed above, scoring functions are limited as they are derived from a test set. Therefore, the score of a given ligand does not tell much unless compared to a reference. In particular cases, the results may be ambiguous, i.e., symmetric ligands (Huang, Grinter & Zou, 2010) and enantiomer pairs (Ramírez & Caballero, 2016). Therefore, the problem is how to obtain quality poses from a scoring function. A first step would be the correct preparation of structures. Notable improvements on docking have been made when molecular dynamics are used to prepare the structures of molecular targets, for instance, to conduct energy minimization and assign charges (Alonso, Bliznyuk & Gready, 2006; Uehara & Tanaka, 2017). For ligand preparation, methods that have improved docking results include: optimization using semi-empiric charges (Adeniyi & Soliman, 2017; Marzaro et al., 2013; Oferkin et al., 2015; Xu & Lill, 2013), COSMO solvation (Oferkin et al., 2015), molecular mechanics Poison-Boltzmann/surface area (Halperin, Wolfson & Nussinov, 2002), and force field optimization (Zoete, Cuendet, Grosdidier & Michielin, 2011). Moreover, it has been shown that forcefield optimization with implicit solvent is comparable to advanced semi-empirical methods (e.g. CHARMM-GBSW vs PM7) and allows for a better energy prediction of complexes (Sulimov, Kutov, Katkova, Ilin & Sulimov, 2017). It also has been suggested that binding may be better predicted by improvement on model representation. To this end, efforts have been conducted to make additional models for binding. Yet, this may not be necessarily the case as very specific descriptors and models do not show significant improvement on the prediction of binding affinity (Ballester, Schreyer & Blundell, 2014).
In addition to modify the scoring functions, some programs allow adjusting the scoring parameters and weight assignment e.g., Vina. Additional refinement includes the consideration of rotational terms and solvation (Kitchen, Decornez, Furr & Bajorath, 2004). However, these processes need to be tested against robust sets like DUD-E or CSAR (Da & Kireev, 2014; Huang, Shoichet, & Irwin, 2006; Huang, Grinter & Zou, 2010).
Current research has turned to machine learning to improve docking and scoring (Waszkowycz, Clark & Gancia, 2011). Machine learning studies the development of adaptive programs capable of self-improvement without user input (Grosan & Abraham, 2011). These techniques may be classified as feature-based which construct vectors, based on a set of instances, and similarity-based which handle explicit input as matrices (Ezzat, Wu, Li & Kwoh, 2017). Common techniques are multiple linear regression, support vector machines, random forests, and neural networks (Ashtawy & Mahapatra, 2014). A significant feature of these methods is their applicability to large sets ( Wang & Wang, 2001).
Approaches that have implemented optimization of scores include PostDOCK which accounts for 3D features that are not reflected on scoring (Knegtel, Kuntz & Oshiro, 1997), Supervised Consensus Scoring using random forest (Bjerrum, 2016), Gradient boosting consensus using decision trees (Ericksen et al., 2017), and several attempts to study the non-linear dependency of ligand orientation (Khamis, Gomaa & Ahmed, 2015). A recent study has used machine learning to identify significant parameters to construct functions with improved prediction of affinity (Bjerrum, 2016).
For pose prediction on the other hand, metaheuristics have been essential for docking algorithms. These techniques are related to artificial intelligence as they are focused on optimization algorithms (Bozorg-Haddad, 2018; López-Camacho, García Godoy, García-Nieto, Nebro & Aldana-Montes, 2015). Examples are simulated annealing, genetic and evolutionary algorithms, ant colony optimization, swarm intelligence, tabu search, and local search. Metaheuristics have been useful for problem solving as they are not prone to combinatorial explosion. Nonetheless it is important to remember that although their algorithms search for optimal solutions, their performance requires problem-specific adaptation (Glover & Sörensen, 2015). Most docking programs have used these methods for pose generation and selection.
Conformational searches on the other hand, are competent enough to sample drug-like molecules. Their apparent underperformance can be attributed to the lack of implicit solvation and, in some cases, to the protonation state as it further deviates the RMSD when compared to crystal structures (Ballester, Schreyer & Blundell, 2014; Gürsoy & Smieško, 2017).
Lastly, docking poses may be ranked based on 3D-similarity against a crystallographic reference. Using this approach better enrichments were found, around 10% better (AUC of 0.7 for docking vs 0.8 for 3D similarity) than scoring rankings (Anighoro & Bajorath, 2016).
Water solvation and docking
Removal of explicit solvent (specifically water molecules) has been a constant criticism in docking (Fischer, Merlitz & Wenzel, 2008). This consideration is often based on the approaches of the scoring function and computational time to limit the degrees of freedom of a given system. Notwithstanding, this approach is out of the question for proteins with highly conserved structural water molecules e.g., HIV-1 protease (Corbeil & Moitessier, 2009) and BET-bromodomains (Crawford et al., 2016; Galdeano & Ciulli, 2016; Zhao, Gartenmann, Dong, Spiliotopoulos & Caflisch, 2014). Nevertheless, the problem is not just limited to the inclusion of water molecules during calculations, but to correctly evaluate water contribution to binding and its implications (Fischer, Merlitz & Wenzel, 2008; Parikh & Kellogg, 2014). First efforts to include water molecules during docking calculations suggested that no significant improvement on scoring was obtained (Roberts & Mancera, 2008). However, some ligands are especially designed to displace water. In such cases, docking simulations are more accurate with the correct treatment of water molecules (Corbeil & Moitessier, 2009).
Currently most docking programs can assess the presence of water molecules during calculations. However, some challenges persist such as scoring reparametrization, pose comparison involving different interacting waters, and correct discrimination of displaceable/non-displaceable waters (Waszkowycz, Clark & Gancia, 2011). Hence, the first step is the identification of key water molecules and their contribution (Kumar & Zhang, 2013).
Methods that have advanced the correct representation of water molecules in proteins include: free energy perturbation methods (Jorgensen & Thomas, 2008), Monte Carlo probability (Parikh & Kellogg, 2014), molecular dynamics of water on the binding site (as implemented by Schrödinger (Kumar & Zhang, 2013; Waszkowycz, Clark & Gancia, 2011), water displacement as implemented by PLANTS (Korb, Stützle & Exner, 2009), “Attachment” of water molecules to ligands as additional torsions (Lie, Thomsen, Pedersen, Schiøtt & Christensen, 2011), QM/MM hybrid methods (Xu & Lill, 2013), COSMO solvation, and semi-empirical charges for ligands (Oferkin et al., 2015). Additional methods are “hydrated docking” scripts for Autodock (Forli et al., 2016), protein-centric and ligand centric hydration as implemented by Rossetta (Lemmon & Meiler, 2013), Water docking using Vina (Ross, Morris & Biggin, 2012; Sridhar et al., 2017), WScore (Murphy et al., 2016), and grid inhomogeneous solvation theory applied by Autodock (Uehara & Tanaka, 2016).
Covalent docking
Covalent inhibitors or modifiers are usually deprioritized as potential drugs candidates due to toxicity concerns. Notable examples are modulators of acetyl-cholinesterase (King & Aaron, 2015). In contrast, instances where covalent ligands are useful have gained notoriety such as Romidepsin, a histone deacetylase inhibitor and prodrug (VanderMolen, McCulloch, Pearce & Oberlies, 2011). Actually, covalent modifiers may be more selective and effective, and more specific for infectious diseases (Schröder et al., 2013). The overall interest towards rational design and development of covalent inhibitors is growing. Current programs for covalent docking include Autodock, CovDock/Glide, DOCK, FITTED, FlexX, GOLD, ICM, and MOE. However, covalent interactions and reactions are not considered explicitly on most programs (Brás, Cerqueira, Sousa, Fernandes & Ramos, 2014). To define the possibility of a covalent interaction, a “link atom” must be defined and manually selected while the docking algorithm forces both parts to occupy the same volume to simulate a covalent bond (Awoonor-Williams, Walsh & Rowley, 2017). The main drawback of current models for covalent interactions is that they just mimic a bond formation between known instances but the process of formation of a covalent bond must be assessed correctly by quantum mechanics (De Cesco et al., 2017). Nonetheless, it is generally accepted that covalent processes are preceded by non-covalent stabilization and orientation of “warhead” groups for reaction (Ai, Yu, Tan, Chai & Liu, 2016).
In addition to mainstream programs, approaches for covalent docking, include CovalentDock a grid based algorithm that uses Autodock scoring and includes bond formation as a new energy contribution, although it is currently limited to certain reactions (Kumalo, Bhakat & Soliman, 2015) and DOCKTITE a collection of SVL scripts for MOE implementation. This approach uses a “warhead screening approach” and chimeric pose generation which can be optimized by force fields (Scholz, Knorr, Hamacher & Schmidt, 2015). Other approach is Steric Clashes Alleviating Receptor (SCAR). This strategy is based on protein mutagenesis to remove reactive residues and favor non-covalent interaction (Ai, Yu, Tan,Chai & Liu, 2016).
Despite the fact that covalent docking remains an unsolved problem, notable progress has been made with current approximations: covalent virtual screening (Toledo Warshaviak, Golan, Borrelli, Zhu & Kalid, 2014), identification of TAK1 inhibitors (Fakhouri et al., 2015), ubiquitin-proteasome pathway inhibitors (Li et al., 2014), and identification of enzymatic substrates (London et al., 2015).
Metal-binding docking
It is well known the high importance of metal cofactors on many protein families. Similar to covalent docking, modeling ligand-metal interactions still requires major developments. In particular modeling interactions with zinc, calcium, and magnesium (which are prominent metal ions in drug discovery).
Correct parametrization of zinc is taking a major attention as it is present on druggable targets and its geometry is mostly tetrahedral (Irwin, Raushel & Shoichet, 2005; Mccall, Huang & Fierke, 2000). Because ligand-zinc interaction may be quasi-covalent its modeling requires different considerations i.e., its acidic behavior allows for “special” hydrogen bonding (Moitessier et al., 2016). This lead to force field reparametrization as implemented by Autodock (Santos-Martins, Forli, Ramos & Olson, 2014), which when compared with other programs yielded a more accurate solution to pose and scoring (Bai et al., 2015; Chen et al., 2007). Programs featuring metal recognition include Autodock, DOCK, FITTED, FlexX, Glide, LigandFit, MOE, and MpSDock.
Conclusions
Molecular docking is a useful technique in structure-based drug design, virtual screening, and optimization of lead compounds. To date, there are several standalone and online docking tools to assist the computational and medicinal chemist to help explain the activity of compounds at the molecular level or to filter compounds for further studies. The increasing number of publications associated with molecular docking reflects the interest of the scientific community to continue developing or refining docking tools, and/or using such tools in drug discovery projects.
Current docking programs are able to predict, in general, correct binding modes. However, a major limitation of most docking protocols is the calculation of accurate binding energies. Such limitations are largely associated with the large number of approximations considering during a docking run such as the treatment of solvent and the flexibility of the macromolecular system e.g., protein-ligand complex. Other major challenges are covalent docking and the accurate and fast modeling of metallic interactions during the docking process.
For practical applications, docking should integrated with other experimental or computational techniques, depending on the primary goals of the study. For instance, biophysical or biochemical experiments have to be done to confirm the putative binding or biological activity predicted with docking. Indeed, virtual screening campaigns are multidisciplinary cycles which involve complementary techniques to further improve hit identification and optimization. Molecular dynamics, free energy perturbation methods and quantum mechanics/molecular mechanics (QM/MM) calculations are instances of methods that can be conducted to improve the calculation of binding energies. These methods are usually applied for selected binding poses generated with docking. In any application, the use of a docking program needs careful selection and validation to correctly assess its prediction power towards a specific problem. Additionally, any preliminary results should be contrasted to at least two more programs or structures using a consensus approach. While some aspects of simulation and calculation still need improvement, current docking methods may provide valuable guidance to drug design and optimization.











 nueva página del texto (beta)
nueva página del texto (beta)



