1 Introduction
In recent years, a deluge of large-scale transcriptomics data from high-throughput sequencing is increasingly raising the demand in computing power and storage. Processing this enormous amount of data requires the use of specialized techniques such as scientific workflow management systems (SWfMSs) and high-performance computing (HPC) resources to extract knowledge from data-intensive RNA-seq experiments [1, 9].
Scientific workflows deal with automating the execution of computational tasks and are needed for improving reproducibility and productivity. They have been used by scientists in a wide variety of domains, including astronomy, bioinformatics, physics, biology, biodiversity, among many others. Scaling workflows on large HPC systems is not an easy task due to the size and nature of data to be processed, the inherent complexity of workflows, the number of workflow instances to be executed, and the complexity of large HPC systems [9].
We identified the following capabilities of SWfMSs relevant to transcriptomics analysis: workflow modularity and automated elasticity to enable checkpointing; scalability concerning the use of the number of workflow tasks versus the number of nodes per run; robustness and fault tolerance due to data issues, resource unavailability, or aborted executions; reproducibility via data provenance recording; portability across computing environments from desktops to parallel and distributed clusters; interoperability of metadata and the use in the same workflow into several programming languages; and ease of development by users with different skill levels in informatics.
In this paper, we present how a collection of performance metrics of well-established transcriptomics software can be orchestrated to optimize the performance behavior of current scientific RNA-Seq workflows. Due to the massive amount of scientific transcriptomics data, the complexity of scientific applications, and the features of distributed computing, the performance analyses require data metrics, information of the workflow execution, and to understand the environmental system as a whole. This also can include capturing the input and experimental provenance data, optimizing the workflow structure, and gathering and storing performance information such as CPU and memory usage and I/O operations at the system level.
Using the Parsl-RNASeq workflow, we are able to execute data-intensive transcriptomics software in HPC infrastructure to enable performance optimization of the workflow execution in a useful way. We have validated our approach by executing a massive amount of cardiomyocyte sequencing data of the evolutionary conserved Wnt pathway, using normal and anomalous conditions. Zelarayan et al. [7] used an in vivo mouse model in which β-catenin is acutely stabilized in adult cardiomyocytes, leading to increased ventricular TCF7L2 expression and activation of target genes. The aim is to understand the consequences of increased Wnt signaling pathway activity, comparing transcriptome profiles of normal (Cre recombinase “positive” control with a WT β-catenin locus) and β-catenin stabilized murine adult cardiac ventricles.
Our main goal is to study the viability of efficiently executing transcriptomics workflows on large HPC systems. The main contributions of this paper include 1. The collection and analysis of performance data from transcriptomics workflows; 2. The case study use of a real-world RNA-Seq workflow; and 3. The analysis of HPC performance metrics as CPU and memory usage, I/O operations, job dependencies, among others.
This paper is organized as follows. Section 2 brings the background on RNA-Seq differential gene expression. Section 3 describes the specification and implementation of the ParslRNA-Seq workflow. Section 4 presents materials and methods. Section 5 shows experimental results. Finally, Section 6 summarizes our findings and discusses future work.
2 Background on RNA-Seq
RNA sequencing (RNA-Seq) uses the capabilities of high-throughput sequencing methods to provide insight into the transcriptome of a cell in a given physiological or developmental condition as diseases derived by genetic variation e.g., cancer. Beyond quantifying gene expression, the data generated by RNA-Seq facilitate the discovery of novel transcripts, identification of alternatively spliced genes, and detection of allele-specific expression. RNA-Seq investigates different populations of RNA including the study of alternative splicing, Single Nucleotide Polymorphisms (SNPs), post-transcriptional modifications, and changes in gene expression over time or between treatment groups or disease progression.
Differential Gene Expression analysis (DGE) allows for elucidating the expression level between different experimental conditions and establishing whether there is a significant difference between them. DGE of RNA-seq data generally consists of three components: normalization of counts, parameter estimation of the statistical model, and tests for differential expression [4].
The data count of samples is tabulated containing the number of sequence fragments assigned to each gene. The quantification and statistical inference are established between systematic changes and variability of different conditions. Testing differential expression, provided in the DESeq2 package, uses negative binomial generalized linear models to estimate dispersion and logarithmic changes and to incorporate prior distributions based on data.
3 ParslRNA-Seq Scientific Workflow
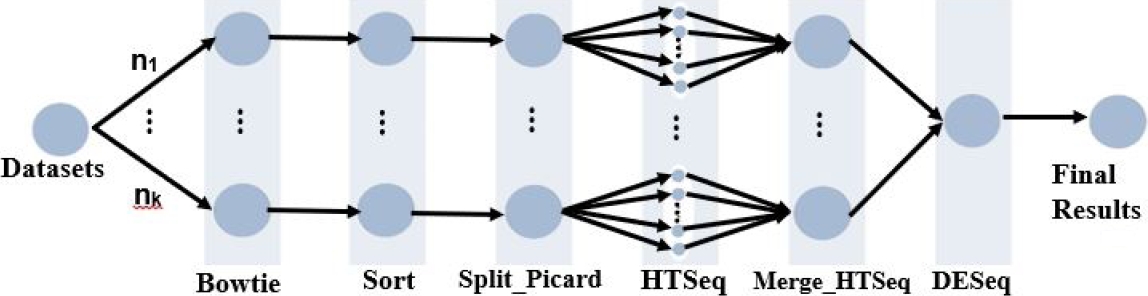
This section presents the conceptual specification of the ParslRNA-Seq workflow for differential gene expression analysis (DGEs), available from https://github.com/lucruzz/rna-seq. ParslRNA-Seq receives four data information: the reference genome of Mus musculus; the gene transfer format (GTF) file used to hold information about gene structure; the FastQ files designed to handle sequence and quality scores represented as single ASCII characters; and a CSV text file containing the list of FastQs and metadata from experimental conditions.
ParslRNA-Seq is composed of six activities (illustrated in Figure 1). Activity 1 executes the Bowtie2 package that maps and compares genome readings, character by character. Activity 2 executes the Samtools program that orders readings and generates a compressed binary format. Activity 3 executes the Picard program to handle and split the readings files into
Activity 4 runs the htseq-count program (HTSeq package), which counts the overlap of reads with genes in DGE. HTSeq sends the mapped reading files to each of the
Parsl [2], a parallel scripting library, provides an easy-to-use model composed of Parsl-Python functions and supports the management and execution of transcriptomics software, assuring reproducibility. Parsl manages the parallel execution of ParslRNA-Seq by applying parameter sweep mechanisms in HPC clusters. Each processing unit operates on the data independently via separate instruction streams. ParslRNA-Seq provenance data is automatically captured by Parsl. Parsl has already been successfully experienced in other complex computing-intensive bioinformatics experiments in HPC environments [5, 6]. Experimental results reinforce the importance of ParslRNA-Seq to help scientists in detecting DEGs from raw sequencing data, with Parsl supporting the management of tasks and provenance data in HPC environments.
4 Materials and Methods
4.1 RNA-Seq Data
Activation of the evolutionarily conserved Wnt pathway has been reported during maladaptive cardiac remodeling. However, the function of Wnt-transcriptional activation in the adult heart is mainly unknown. Zelarayan et al. [7] performed the transcriptome and genome analysis at the University Medical Center, Goettingen. RNA was isolated from mice cardiac tissue and RNA libraries were prepared for sequencing using standard Illumina protocols. Sequence reads were aligned to the mouse reference assembly (UCSC version mm9) using Bowtie 2.0. For each gene, the number of mapped reads was counted using htseq-count and DESeq2 was used to analyze the differential expression. Mus musculus GEO.ID is GSE97763.
This study belongs to a real RNA-Seqfn experiment. It uses six FastQ sequencing data files of cardiac ventricles deposited in Gene Expression Omnibusfn (GEO) public repository under accession GSE97763 and GSE97762 for RNA-seq datasets. FastQs accession numbers of the control condition group are: SRR5445794, SRR5445795, SRR5445796 and for the Wnt pathway: SRR5445797, SRR5445798, SRR5445799, including datasets of varying sizes (1.8 to 3 GB).
4.2 Experiment Setup
We follow the same protocol adopted by Zelarayan et al. 2018 [7] to validate and analyze transcriptomics results obtained by the executions performed with the ParslRNA-Seq scientific workflow. The transcriptomics software used in experiments are Bowtie2fn program, Samtoolsfn program version 1.10, Picardfn program version 2.25.0, HTSeqfn framework version 0.13.5 with the htseq-count script, HTSeq-Merge Python homemade-script, and DESeq2fn package. All software, libraries and dependencies, Parsl and Python components, Intel VTune Profilerfn, and Darshanfn tool were deployed at the top of the Santos Dumont environment.
4.3 Santos Dumont Supercomputer
The Santos Dumont (SDumont) supercomputer, one of the largest in Latin America, is located at the National Laboratory for Scientific Computing (LNCC/MCTI, Brazil). SDumont has an installed processing capacity of 5.1 Petaflops with a total of 36,472 CPU cores distributed across 1,134 compute nodes. SDumont has a Lustre parallel file system, integrated into the Infiniband network, with a raw storage capacity of around 1.7 PBytes and a secondary file system with a raw capacity of 640 TBytes. For our experiments, six nodes of SDumont were utilized, each node uses two Ivy Bridge Intel Xeon E5-2695v2 CPUs (12c @2.4GHz) and 64 GB of RAM. For more information, visit http://sdumont.lncc.br.
4.4 Parsing RNA-seq Files Strategy
The NGS’s big challenge lies in the enormous data size for every single sample analyzed. A strategy to optimize parallelization in an HPC environment is to split input data into small and equally-sized portions. For parallelized read mapping, the alignment is carried out in parallel either by making use of array jobs or by distributing the data across multiple threads using OpenMP or across multiple compute nodes using the message passing interface (MPI).
We propose strategies managed by Parsl that scale to hundreds of threads better than single processed workflows or pipelined approaches. We explore how the FastQ file format, its unpredictable record boundaries, in particular, can impede thread scaling. We suggest a way to modify FastQ files while dividing the file size into blocks and how including these activities in workflow enables further improvements in thread scaling.
In RNA-Seq, to improve thread scaling we should restructure inputs and outputs, converting standard FastQ (or SAM) files to blocked FastQ files [8], where the number of input reads per block (N) are 24, to make best use of the 24 threads per node used in SDumont. The number of FastQ reads per thread must be chosen for each ParslRNA-Seq configuration and system.
ParslRNA-Seq starts with Bowtie (first activity); then the second activity Sort sorts the SAM or BAM files. The third activity Split Picard retains the sort order of reads matching to the original BAM, splits files, separates files into Nth reads, and finally creates an output directory for storing split BAM files. In the fourth activity, HTSeq processes split files by calling the “–nprocesses = 24” argument.
4.5 Multithreading & Multiprocessing Strategy
We consider in this work that a Multiprocessor (MP) is a system with more than one processor that assigns separate memory and resources for each process. Conversely, Multithreading (MT) is a programming model in which multiple threads run collaboratively in a single processor. Creating multiple threads inside a single process often help increasing performance. Moreover, we notice that modifications in some ParslRNA-Seq activities (mainly Bowtie2 and HTseq) are potential points to explore MT or MP thread scaling, as they can increase the computing speed of the system.
The ParslRNA-Seq workflow code (https://github.com/lucruzz/RNA-seq/blob/master/RNA-seq.py) shows the software command lines. While Bowtie2 creates MT processes, HTSeq “–nprocesses” (MP argument) only works to process different BAM files in parallel, i.e., htseq-count on one file is not parallelized. For instance, let us consider the following context in our ParslRNA-Seq processes. While we focus on making the best use of threads in a single process, an alternative is to run multiple simultaneous processes, possibly with many threads each. ParslRNA-Seq consumes six input FastQs, each deployed in parallel in an independent node.
For each node, Bowtie2 sets the performance option “-p/–threads NTHREADS” to launch the number of parallel search threads (default: 1) to process each FastQ. The threads will run on separate processors/cores and synchronize when parsing reads the output alignments, increasing alignment throughput by approximately a multiple of the number of the threads (linearly).
Split_Picard “SplitSamByNumberOfReads” option splits an input query-grouped SAM or BAM file into multiple (e.g., 24) BAM files while maintaining the sort order to parallelize alignment. The HTSeq “–nprocesses” processes those BAM files.
MP can suffer from load imbalance as some batches take longer to execute than others, and the job’s duration is determined by the longest-running batch. Merge HTSeq suffers this impact whereby some lock-holding threads are slow to finish their works (and release the lock) due waiting threads are using its resources. Finally, DESeq2 should wait for Merge HTSeq finishes to be executed.
4.6 Profiling Strategy
A global overview of the CPU and I/O systems is the first step to understanding the computational demands on the machine and detecting opportunities to optimize application performance, system performance, and system configuration for HPC.
We obtained and studied CPU and I/O performance reports obtained on the Santos Dumont supercomputer (SDumont). Those were obtained with the Intel VTune and the Darshanfn I/O profiling tools, for CPU and I/O, respectively. This choice was motivated by a previous study of SDumont’s performance conducted by Bez et al., 2020 [3], which provides details about how to characterize the application I/O phases from coarse-grained aggregated traces using Darshan.
Intel VTune provides insight into CPU and threads performance, scalability, bandwidth, caching, and more. VTune collects the tallies from all the cores’ counters at frequent intervals; when the run is over, it analyzes the collection and presents the results via a GUI. The Darshan tool version 3.1.4 profiles executions on I/O metrics. Bowtie2 and HTseq are the most representative CPU and time-consuming software of ParslRNA-Seq executed in SDumont in 2021, and they were the main focus in our case studies.
5 Results and Discussions
In this section, we present the performance and scalability of the parallel executions of transcriptomics scientific workflows. We have evaluated the CPU and I/O behavior of the ParslRNA-Seq executions in the Santos Dumont supercomputer. The CPU analyses are extensions of [5, 3].
5.1 CPU Performance Results Using VTune
ParslRNA-Seq CPU Performance. ParslRNA-Seq allocates, executes, and manages each of the six FastQs in one node of 24 threads. CPU and I/O influence the HPC scalability of the workflow. The original workflow modeling -of three activities- ([5, 6]) has been modified to optimize parallel and distributed executions and improve the total execution time (TET) (Figure 2, Figure 3, and Figure 4). The actual ParslRNA-Seq workflow — of six activities — was described in Figure 1.

Fig. 3 ParslRNA-Seq multithreading performance (seconds) on a single node. The y axis is different for each plot
Figure 2 presents the TET achieved by each of the six activities of ParslRNA-Seq. Bowtie2, Split Picard, and HTSeq are the activities with the longest execution times (Figure 2), while Bowtie2 and Sort are the most I/O-intensive activities, i.e., they spend more time computing I/O relative to their total execution time (Figure 5). Then, we focused on parsing files, multithreading, and multiprocessing to improve the workflow behavior, mainly for the Bowtie2 and HTSeq activities.
ParslRNA-Seq Multithreading Performance. Figure 3 presents the workflow efficiency for the multithreading performance. The figure plots the node count on the horizontal axis and maximum per-thread wall-clock time, i.e., the time required to align all reads, on the vertical axis. The figure shows both versions of ParslRNA-Seq with (a) the previous version of three activities and (b) the ParslRNA-Seq modified model of six activities. We can see that the new model, while adding activities, improved performance and efficiency.
In Figure 3(a) Bowtie2 multithreading executes a task for each (not parsed) FastQ file in a node (24 threads). In addition, HTSeq executes each (not parsed) file in an entire node, i.e., no MT or MP strategy was applied. In Figure 3(b), Bowtie2 multithreading also executes a task for each (not parsed) FastQ in a node. Nevertheless, HTSeq executes a task of each SAM block in a thread (a SAM file was parsed in 24 blocks), i.e., each block is assigned to a thread.
A better distribution in the use of CPU cores in parallel was observed in Figure 3(b) due to the use of MP and MT approaches. However, that also required the insertion of extra activities in the workflow modeling. So despite the parallel execution of tasks performed by Parsl, there is still a considerable number of idle CPUs in most of the workflow execution. Figure 3(b) shows the use of up to 20 CPUs has an effective average CPU utilization considered as “poor” but over 20 CPUs the simultaneous use of processors presented an “ideal” utilization.
ParslRNA-Seq Multiprocessing Performance. Figure 4 shows two multiprocessing scenarios executed on SDumont: previous ParslRNA-Seq version of three activities in red and the optimized ParslRNA-Seq version of six activities in blue. With a single node, the new ParslRNA-Seq version (in blue) improves performance over the previous version from 72 to 65 minutes.
We further increased the number of nodes from 1 to 6 (with 24 cores per node).
This experiment consumed six FastQs by execution. The previous workflow version does not scale with the number of nodes (the execution time remains the same). On the other hand, the optimized workflow’s performance is improved with more available nodes and cores. Time was decreased from 65 minutes (using 24 cores) to 24 minutes (using 144 cores), which means a speed-up (an acceleration factor) of
5.2 I/O Performance Results Using Darshan
The workflow was executed with the Darshan profiler to investigate its I/O behavior. For discussion, we analyzed two input sizes,
For both applications, increasing the input size increases the proportion of the execution time spent on I/O. That indicates that the I/O limits the scalability of these codes: as more data is treated, most time is spent on I/O and thus the CPU-focused optimizations presented earlier may have less impact on performance.
I/O Analysis for Bowtie2. Changing the input from
I/O Analysis for Sort. Time increased from 41 seconds (
5.3 Biological Analysis
Our biological performance results were validated to the reported by Zelarayan et al. [7] at the Gottingen University, in a collaboration between our research groups, reporting almost identical biological results. In the present article, we proposed the ParslRNA-Seq workflow and the computational executions performed in SDumont.
6 Conclusions and Future Work
In this work, we have presented a real-world workflow analysis for data-intensive transcriptomics applications to enable performance optimization of HPC systems. To this end, we have evaluated various transcriptomics applications from ParslRNA-Seq to analyze massive amounts of RNA-seq data in a controlled environment. We have used Parsl as a workflow management system, VTune and Darshan as profiling tools, and the SDumont as our machine. Moreover, we have developed a new ParslRNA-Seq version tailored to the needs of tracking a workflow execution and identifying potential issues to improve performance.
Our experiments demonstrate that this optimized workflow can accurately orchestrate computation resources, helping to pinpoint relevant metrics to help identify performance problems. Our results show performance improvements of up to 63.08% of ParslRNA-Seq executions, from 65 minutes (24 cores) to 24 minutes (144 cores). Additionally, we characterized the I/O behavior of the workflow components, identifying I/O problems in two of them, which will be the focus of future optimization efforts.
Indeed, we plan on continuously improving the ParslRNA-Seq modeling and performance. We want to explore the possibility to stage intermediate data on computing nodes to minimize parallel file system activity. Furthermore, we want to include system-level monitoring data in our analysis, which may explain the observed behaviors, particularly regarding I/O. Finally, we plan to provide a mechanism to track data and metadata to enable offline analysis. We aim to introduce a new database automatically populated by Parsl or another SWfMSs so that users can retrieve workflow performance data. The data collected is a valuable training resource for automated machine learning analysis.











 nueva página del texto (beta)
nueva página del texto (beta)