1 Introduction
Currently, cancer is a worldwide public health problem, is the world’s second leading cause of death, the second cause of death in the United States [22], and the third in Mexico [5]. Electronic Health Records (EHR) contains medical history, diagnoses, medications, laboratory and test results, and treatment plans.
EHR information systems are constantly growing in volume and variety, being an opportunity to carry out data analytics with the different types of structured and unstructured data contained in these records. EHR data analytics has been applied to cancer classification and cancer prediction, obtaining promising results based on correct classification and prediction, respectively. However, data used has been highly filtered for these purposes and data contained in EHR systems is heterogeneous (according to the data type and the data source).
There are new challenges to develop computer models that can work with different types of data (structured or unstructured), and the variety of data (data source) in order to capture as much information as possible to obtain cancer classification models.
In this paper, a modeling process is proposed in order to train three machine learning models for cancer type classification (Support Vector Machines (SVM), Multi Layer Perceptron (MLP), and Adaboost-SAMME). Classification is done for lung, breast and liver cancer types. The aim is to obtain the best performing machine learning model, based on well-known metrics for data analytics.
The model use structured data (i.e. lab tests and demographics), and unstructured data (i.e. free text clinical notes) from the MIMIC-II clinical database by using machine and deep learning techniques. In particular, the Paragraph Vector algorithm is used as a deep learning approach to text representation.
The rest of this paper is organized as follows. Section 2 explores related work associated with machine learning models for cancer classification using structured and/or unstructured data. Section 3 presents the proposed modeling process for cancer type classification using both structured and unstructured data representation. Section 4 presents the experiment and the results obtained from the machine learning models trained on different data configurations (structured, unstructured and both types of data). Finally, section 5 presents the conclusion of this paper.
2 Related Work
Machine Learning models have been applied to cancer classification using structured data. Authors in [14] applied machine learning to breast cancer classification and reported a new feature selection algorithm using structured data, obtaining promising results. In [20], the authors tested the performance of three machine learning algorithms, finding the K-Nearest Neighborhoods (KNN) as the best performing model with a precision of 98.27.
In [24], the authors compared six machine learning algorithms applied to the breast cancer (metastasis survival rate) using structured data, finding the SVM as the best performing model.
In another work [4], the authors compared three machine learning models to help in early detection of breast cancer using a specialized dataset, obtaining that the best performing model for early diagnosis is Random Forests (RF).
In [3], the authors compared machine learning algorithms for lung cancer detection and found the Gradient Boosted Tree as the best performing model, with a precision of 87.82%.
Machine learning models have also been applied to cancer classification using unstructured data.
In [18], the authors proposed a Convolutional Neural Network (CNN) based model for image retrieval of lung nodules, obtaining a precision of 0.73.
In [13], the authors consdiered a model to extract relations from clinical text, then applied Recurrent Neural Networks (RNN), obtaining an improvement of 3% over a baseline model.
In [9], the authors also extracted relations from clinical text, applied Convolutional Neural Network (CNN), and obtaining an average precision of 73.4% on their best performing model.
In [11], the extracted information from dead certificates to obtain statistics for common and rare cancer deaths; they found a combination of rule-based classifiers and SVM as their best performing model.
[6] proposed a system for automatic classification of radiologic reports using machine learning models, obtaining the RF model as the best performing model.
Deep learning-based techniques using unstructured EHR data has shown promising results. In [1], the authors examine the strength of deep learning approaches for pathology detection in chest radiograph data. Convolutional Neural Networks (CNN) are used for identifying different types of pathologies in chest x-ray images. The authors have trained a CNN with ImageNet, a large-scale nonmedical image database, using low-level visual features derived by the concatenation of orientation, color, and intensity histograms over different scales and cell segmentation. Authors obtained an area under curve of 79% for classification between healthy and abnormal chest x-ray.
In [17], a Long Short-Term Memory (LSTM) based neural network called DeepCare is proposed for disease progression modeling, intervention recommendation, and future risk prediction for diabetes and mental health. DeepCare is an end-to-end deep dynamic neural network that reads medical records, stores previous illness history, infers and predicts future medical outcomes by depicting. DeepCare uses word embedding to represent the semantics of diagnoses, interventions, and admissions notes to infer experiences pooled to reason about the current illness states and the future prognosis. Furhtermore, [18] uses a CNN to construct a content based medical image retrieval system for pulmonary nodules. They proposed a UNet method to preprocessing images under the guidance of medical knowledge. Then, a CNN module extract features of the segmented images with different sizes. UNet is considered a deep learning framework that is modeled and trained on a collection of medical images. The features learned by UNet are used to present a highly efficient medical image retrieval system that works for an extensive collection of multimodal datasets. Finally, UNet is modified to learn domain-specific image representations and simultaneously set hash-like (or binary-coded) functions.Their method achieves 73% precision for image retrieval.
Unstructured EHR data need to be represented in vector space known as word embedding. This technique is used to encode words in a space that is subsequently used as input for many machine or deep learning models. Word representation using a distributional semantics of words is addressed in [21], where a comparison of the traditional word embedding methods (word2vec, GloVe, fastText) is presented to extract clinical concepts. They also analyze the impact of the pretraining time of a large language model like ELMo or BERT on the extraction performance. The authors also present an intuitive way to understand the semantic information encoded by contextual embeddings for concept extraction tasks.
Open-domain embeddings and pretrained clinical embeddings from MIMIC-III (Medical Information Mart for Intensive Care III) are evaluated for extracting clinical concepts. [25] proposes a pretrain deep embedding models (BERT) on medical notes from the MIMIC-III hospital dataset. The authors identify dangerous latent relationships that are captured by the contextual word embeddings for clinical prediction tasks that include detection of acute and chronic conditions. [23] adapts datasets about biomedical literature in Spanish, in particular, a considerable volume of EHRs in Spanish. The authors create an in-domain medical word embeddings using FastText model for named entity recognition task.
Existing solutions focus generally on classification using only structured data ([14, 20, 24, 4, 3] applying machine learning models) or using only unstructured data ([18, 13, 9, 11, 6] applying machine learning models and [1, 17, 18] applying deep learning techniques). The novelty of the approach proposed is a modeling process using both structured and unstructured data representation to classify breast, lung, and liver cancer. In particular, this approach use a deep learning approach to represent unstructured data using the Paragraph Vector algorithm and to evaluate the performance of SVM, MLP, and Adaboost models using only structured data, only unstructured data, and using structured and unstructured data.
3 Proposed Approach
This paper proposes a modeling process for breast, lung and liver cancer classification, using structured and unstructured data (see Figure 1). The proposed approach was tested on MIMIC-II clinical database [19]. This database has patient clinical notes as free-text documents and patient structured data (see Figure 1). Patient structured data consist of 19 variables shown in Table 1.
Table 1 MIMIC-II database variables
| Variable | Description | Type A-Alphanumeric N-Numeric |
| SEX | Sex: F or M | Alphanumeric |
| Marital Status | Marital status: single or married | Alphanumeric |
| Ethnicity | Ethnic origin: White, Hispanic or Latino, Asian, Black/African American, etc. | Alphanumeric |
| Religion | Religion: Catholic, Buddhist, Jewish, etc. | Alphanumeric |
| Admission Type | Admission type reason: emergency, elective or urgent | Alphanumeric |
| Admission Source | Admission source: emergency, transfer from hospital, clinic or physical referral | Alphanumeric |
| Height | Patient height | Numeric |
| Weight | Patient weight | Numeric |
| UREA N | Urea nitrogen [Mass/volume] in Serum or Plasma | Numeric |
| PLT CNT | Platelets [#/volume] in Blood | Numeric |
| HCT | Hematocrit [Volume Fraction] of Blood | Numeric |
| HGB | Hemoglobin [Mass/volume] in Blood | Numeric |
| MCHC | Erythrocyte mean corpuscular hemoglobin concentration [Mass/volume] | Numeric |
| MCH | Erythrocyte mean corpuscular hemoglobin [Entitic mass] | Numeric |
| MCV | Erythrocyte mean corpuscular volume [Entitic volume] | Numeric |
| RBC | Erythrocytes [#/volume] in Blood | Numeric |
| CREAT | Creatinine [Mass/volume] in Serum or Plasma | Numeric |
| RDW | Erythrocyte distribution width [Ratio] | Numeric |
| WBC | Leukocytes [#/volume] in Blood | Numeric |
An example of a patient clinical note is in Figure 2. Each patient has a ICD-9 code, an international statistical identifier that classifies diseases and related health problems. A total of 225 patients (75 breast, 75 liver, and 75 lung cancer patients) was used as a balanced set. A total of 10,518 clinical notes (2157 breast, 3653 lung, and 4708 liver cancer notes) were extracted.
The modeling process has two workflows. One workflow extracts patient structured data (see 3.1) and the other extracts patient notes (see 3.2).
Each workflow has a pre-processing phase (structured and unstructured data pre-processing), and alphanumeric data is transformed to its numeric representation. Information obtained from both workflows is used as input values for machine learning models (see 3.3), in order to obtain the best performance model for cancer classification.
3.1 Structured Data Workflow
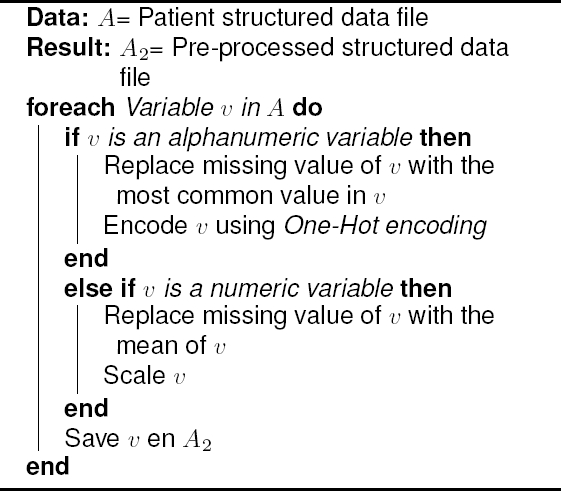
The goal of this structured data workflow is to obtain a tidy patient structured data set, used as inputs for the machine learning models. Algorithm 1 shows structured data pre-processing.
3.2 Unstructured Data Workflow with Deep Learning
The goal of this unstructured data workflow is to obtain a tidy and numeric data representation of patient notes. In this workflow, Natural Language Processing techniques are considered.The first step is text pre-processing in order to prepare each clinical note into a standardized text representation for posterior manipulation. Algorithm 2 shows this step.
In the preprocessing of unstructured data, several task are involved as follows:
— Segmentation. This task is in charge of obtaining the lexical elements of each sentence from clinical notes. First, paragraphs, then sentences, and finally words are obtained.
— Remove words and characters. Stopwords obtained form NLTK library are removed of texts. Words that do not add value to the clinical notes. Also, special characters are eliminated, such as: $ ! ¿ ? + *.
— Word lemmatization. This process aims to to reduce inflectional forms by obtaining the common lemma of each word. The base form is a word when all affixes has been removed. The Snowball Lemmatization with NLTK in Python was used.
The second step is to obtain a Vector Space Representation, that is the process to represent the pre-processed unstructured data as a fixed-length vector of real numbers. The Paragraph Vector (PV) algorithm [12] based on deep learning was chose to got a vector representation for each clinical note. Algorithm 3 shows this transformation. The Paragraph Vector (PV) algorithm is considered within the Deep Learning field since it represents the texts of the clinical notes by a semantic distribution of the words at the paragraph level, capturing the contextual knowledge.
Vector Space Representation Paragraph Vectors (doc2vec) [12] is based on the word2vec [15] algorithm to train and infer word vectors and can be considered a deep learning approach to text representation, because of the use of a neural network to encode text as numerical data. As in the case of doc2vec, the same approach to train the word vectors is taken, but an additional matrix is added, and each column represent the paragraph (document) vector of each document. This algorithm can be divided in two phases, training and prediction phases. The objective in the training phase is to train the word vectors using a neural network, to predicting the context words. The training is done with stochastic gradient descent, in which each step, a fixed-length context words is taken from a random paragraph. Formally, given a set of training words w1, w2,..., wT , the objective is to maximize the average log probability (Eq. 1):
In the prediction phase, a multiclass classification is done, with a softmax classifier as suggested by the authors, as shown in (2):
In equation (2) the yi corresponds to a un-normalized log-probability for each word i obtained with (3):
In equation (3), U, b are softmax parameters, h is constructed by the average or concatenation of paragraph matrix D and word vector matrix W.
Once the model is used to obtain the vector for each patient notes and along with the features from the structured data workflow, a classification using machine learning models can be done.
3.3 Machine Learning Models
Three machine learning models: Support Vector Machines (SVM) [2], Multi-Layer Perceptron (MLP) [16], and Adaboost-SAMME [7] were applied to classify liver, breast, and lung cancer. The algorithms selected for the present work take into consideration some of those used in the related work. The SVM algorithm demonstrates stable performance and, in most cases, one of the best algorithms to carry out the classification; in the same way, the integration of MLP is considered, since recently good results have been obtained in the classification considering methods based on neural networks. The case of AdaBoost is considered given that it is an algorithm that has presented good results and it is also considered as a combination of classifiers to give a final result.
In all the models, a partition of 80% of the data was considered for training and 20% of the data for final evaluation. The implementation of each of the ML algorithms was used using the Python scikit-learn API.
For training the models, it was important to consider the overfitting problem [8], i.e., a model would have a perfect score with the training data but in the case of new datasets, which have never been fed into the model, the model would fail and prediction will be highly affected.
For solving the overfitting problem, a cross-validation process [10] was applied, using the k-fold method. This method split the training set into k smaller sets called folds.
For each of the k folds, the model selects a fold for validation and the others for training. In each iteration, the division of the test set is done differently and is calculated the mean score and the standard deviation of the model.
3.3.1 Score and Model Evaluation
To obtain the best performing model for classification, three standard performance scores were used for multiclass classification: precision, recall and f1-score. Each measure formula can be seen in equations 4, 5, 6, respectively:
where TP, FP and FN stands for true positive, false positive, and false negative, respectively.
4 Experiments and Results
Three experiments were done to evaluate the performance of SVM, MLP and Adaboost-SAMME to classify breast, lung, and liver cancer. The following parameters were configured for each model:
— Parameters for SVM. SVM model was tested with a C value from 1 to 1000 incremented by 100; a γ value from 0.1 to 1 incremented by 0.1; and the kernel RBF.
— Parameters for MLP. MLP model was tested with a hidden layer size from 300 to 600 incremented by 100; the activation functions: identity, logistic, tanh, and relu; the solvers for weight optimization: lbfgs, sgd, and adam; and a maximum number of iterations from 100 to 1000 incremented by 100.
— Parameters for Adaboost. Adaboost model was tested with a learning rate from 0.1 to 1 incremented by 0.01; a maximum number of estimators from 50 to 100 incremented by 1; and using the algorithms: SAMME and SAMME.R.
The first experiment (see 4.1) used only the data obtained from the unstructured data workflow for training and evaluating the machine learning models, the second (see 4.2) used only the data obtained from the structured data workflow, and third (see 4.3) used data obtained from the unstructured and structured workflow.
4.1 Results for Cancer Classification using Unstructured Data
The best performing SVM model using unstructured data had a C value of 100 and γ value of 0.8. Table 2 shows an average precision of 87% for the SVM model. There is a 92% of precision for breast cancer, there is only 8% of breast cancer cases that can be liver or lung cancer. Thus, the recall value is one for liver cancer, meaning that all the relevant breast cancer cases are detected, but with a 87% of precision.
Table 2 Best SVM, MLP and Adaboost models found for cancer type classification with unstructured data
| Cancer type | SVM | MLP | Adaboost | ||||||
| Precision | Recall | F1-Score | Precision | Recall | F1-Score | Precision | Recall | F1-Score | |
| Breast | 0.92 | 0.80 | 0.86 | 0.73 | 1.00 | 0.85 | 0.91 | 0.71 | 0.80 |
| Liver | 0.87 | 1.0 | 0.93 | 0.95 | 0.90 | 0.93 | 0.58 | 0.85 | 0.69 |
| Lung | 0.83 | 0.83 | 0.83 | 0.91 | 0.71 | 0.80 | 0.73 | 0.61 | 0.67 |
| Average | 0.87 | 0.87 | 0.87 | 0.89 | 0.87 | 0.87 | 0.74 | 0.72 | 0.72 |
The best performing MLP model using unstructured data applied the activation function Relu, a hidden layer size of 300, and a maximum number of iterations of 500. Table 2 shows that the MLP model obtained an average precision of 89%. For breast cancer, the recall value is one, meaning that 100% of the relevant cases of breast cancer are detected (there are not falses negatives); however, there are 27% of falses positives (because precision is 73%), meaning that the algorithm classifies as breast cancer but can be liver o lung cancer. It can be observed that this model outperforms the SVM model in precision for liver and lung cancer.
The best Adaboost model using unstructured data applied the SAMME algorithm, had a learning rate of 0.939, and a maximum number of estimators of 90. Table 2 shows an average precision of 74% for the Adaboost model, the worst performing model using unstructured data for liver and lung cancer classification. However, breast cancer obtained a 91% of precision and F1 score of 80%.
4.2 Results for Cancer Classification using Structured Data
The best performing SVM model had a C value of 300 and a γ value of 0.2. Table 3 shows that precision, recall and f1-score has obtained an average just above the 50% for the SVM model. However, breast cancer obtained a 70% of precision.
Table 3 Best SVM, MLP and Adaboost models found for cancer classification using structured data
| Cancer type | SVM | MLP | Adaboost | ||||||
| Precision | Recall | F1-Score | Precision | Recall | F1-Score | Precision | Recall | F1-Score | |
| Breast | 0.70 | 0.74 | 0.72 | 0.44 | 0.67 | 0.53 | 0.63 | 0.67 | 0.65 |
| Liver | 0.56 | 0.38 | 0.45 | 0.65 | 0.65 | 0.65 | 0.46 | 0.46 | 0.46 |
| Lung | 0.44 | 0.54 | 0.48 | 0.60 | 0.38 | 0.46 | 0.46 | 0.43 | 0.44 |
| Average | 0.56 | 0.55 | 0.55 | 0.56 | 0.56 | 0.56 | 0.52 | 0.52 | 0.52 |
The best performing MLP model used the activation function identity, a hidden layer size of 300, and a maximum number of iterations of 500. Table 3 shows a 65% and 60% of precision for liver and lung cancer, respectively. MLP model performs better than the SVM model. The results show more consistent precision values than with the SVM model, having a precision above 60% for liver and lung cancer, but breast cancer precision is bad.
The best Adaboost model used the SAMME algorithm, had a learning rate of 0.25, and a maximum number of estimators of 65. Table 3 shows a poor precision performance for liver and lung cancer classification (46%) compared to the SVM and MLP models.
4.3 Results for Cancer Classification using Structured and Unstructured Data
The best performing SVM model using structured and unstructured data had a C value of 100 and γ value of 0.1. Table 4 shows an average precision of 84%, breast cancer has a 93% of precision, liver cancer obtained a 92% of precision and lung cancer a 69% of precision. Precision was better than with only unstructured data. In fact, precision values for breast and liver cancer excel and surpass the unstructured model, but for lung cancer shows poor performance. For lung and liver cancer, recall is high but shows poor performance for breast cancer.
Table 4 Best SVM, MLP and Adaboost models found for cancer type classification with both structured and unstructured data
| Cancer type | SVM | MLP | Adaboost | ||||||
| Precision | Recall | F1-Score | Precision | Recall | F1-Score | Precision | Recall | F1-Score | |
| Breast | 0.93 | 0.68 | 0.79 | 0.94 | 0.89 | 0.92 | 0.80 | 0.75 | 0.77 |
| Liver | 0.92 | 0.92 | 0.92 | 0.64 | 0.75 | 0.69 | 1.0 | 0.83 | 0.91 |
| Lung | 0.69 | 0.92 | 0.77 | 0.77 | 0.71 | 0.74 | 0.70 | 0.82 | 0.76 |
| Average | 0.84 | 0.84 | 0.83 | 0.80 | 0.80 | 0.80 | 0.83 | 0.80 | 0.81 |
The best performing MLP model using structured and unstructured data applied the activation function Logistic, a hidden layer size of 400, and a maximum number of iterations of 700. Table 4 shows an average precision of 0.80, breast cancer has a 94% of precision, better than the results found using only unstructured data. However, precision for liver and lung cancer dropped compared to the unstructured MLP model.
The best Adaboost model using structured and unstructured data applied the SAMME algorithm, had a learning rate of 0.369, and a maximum number of estimators of 55. Table 4 shows an average precision of 83%. For cancer liver classification, precision value is one, meaning that all the liver cancer cases are detected (there is no false positives); however, there is a 83% of recall, meaning that there are falses negatives (there are liver cancer cases that are not detected).
5 Conclusion
This paper has proposed a modeling process for breast, lung and liver cancer classification using structured and unstructured data. A deep learning approach has been useful to represent unstructured data (clinical notes) using the Paragraph Vector algorithm. Three experiments were made to evaluate the performance of SVM, MLP, and Adaboost models: using only structured data, only unstructured data, and using structured and unstructured data.
Results showed that precision was better for the models using just unstructured data, having stable scoring across all cancer types, the MLP model is the best model for liver and lung cancer, and SVM model for breast cancer. The results from the experiment using only structured data are poor (average scores are just above 50%). In the third experiment, using structured and unstructured data, an improvement for some scores is observed, mainly for the SVM and Adaboost models, although this small improvement does not justify the fall in performance for the other cancer types for the same models.
Finally, based on the consistent results and stable scores, the best performing model is MLP trained with unstructured data, achieving 89% of precision.
The main contributions of this paper are a) a structured and unstructured data combination approach to the classification of Electronic Health Records of cancer; b) Natural Language Processing techniques for Unstructured Data (Clinical Notes) from Electronic Health Records of cancer patient; c) a comparison of three machine learning algorithms for classifying the records; d) the application of a deep learning technique for the representation of texts from clinical notes.
The use of Deep Learning for the distributional semantic representation of words at the paragraph level is a feature that makes this paper an outstanding one on existing works in the state of the art since its techniques are applied in all experimental setups. This application makes the traditional ML algorithms improve, achieving promising results.
As future work, data can be obtained from specific cancer studies to check whether this kind of filtered data helps to improve the unstructured data model in a better way than the general patient data did. Also, it is worth to test this workflow using CNN models.











 nueva página del texto (beta)
nueva página del texto (beta)