1 Introduction
Modern healthcare applications are largely aided by Clinical Decision Support Systems (CDSSs) which mostly involve predictive and preventive analytics applications for betterment of patient healthcare delivery. Digital revolution has led to continuous generation and streaming of huge amount of data in the form medical records, radiology reports, prescriptions, clinical notes, scan images, etc. The continuing research in developing CDSS by making use of the huge amount of generated medical data indicate the significant efforts to augment the decision making made by medical caregivers.
Several Machine Learning (ML) and Data Mining based CDSSs have been developed over the years, helping caregivers make informed decisions and timely interventions during critical conditions of patients. Some major CDSS applications like mortality prediction [5,4,8,16], hospital readmission prediction [13], length of stay prediction [19,1], disease-specific prediction [9,10,20], generic disease or iCd91 diseases/group prediction [17,15,6] and disease coding of discharge summaries [2,3,7,21] have gathered significant research interest in the field of healthcare and biomedicine.
Most of the existing CDSSs developed over the years largely depend on the availability of structured patient data in the form of Electronic Medical Records (EMRs) or Electronic Health Records (EHRs), which is suited for usage in developed countries due to the large scale adoption of EHRs in hospitals. However, hospitals in developing countries still depend on clinical notes of free and unstructured text format. Therefore, there is a crucial need for alternative methods to develop effective CDSSs without relying on structured hospital or patient data.
Purushotham et al. [17] benchmarked three prediction models - ICU mortality, ICD9 group prediction and hospital readmission on large healthcare data using Super Learner and Deep Learning techniques. The ICD9 group prediction task presented in their work is a generic disease prediction task that can be a good CDSS for caregivers and can also be considered as a prior step for ICD9 code prediction.
This work was also based on structured patient data and the benchmarking was performed on various feature subsets and the best performing model used 105 features which included a variety of feature values such as input events (fluids and medications given through IV), output events (urinary and rectal output), prescribed medicines and other ICU events like chartevents (readings noted during ICU stay) and labevents (lab test results).
As deep learning models can learn to classify or regress from numerous raw feature values quite well, their approach achieved good results. However, in a real world scenario, to measure all these values, convert them into a structured EHR form and then to predict an outcome, a significant time delay is inevitable, which might lead to worsening of patient condition. Thus, disease prediction models that can make predictions with high accuracy with with low latency, which can predict with high accuracy over lower test data are the need of the hour.
In this paper, a feature modeling approach that adopts the concepts of word embedding and ontologies to model features effectively to train and build a neural network based model to predict diseases (ICD9 disease group) is presented. We show the results of the benchmarking study of our proposed model (modeled on unstructured radiology notes) against the current state-of-the-art model (built on structured clinical data), where our model performed on par with the state-of-the-art model.
The rest of this paper is structured as follows: Section 2 describes the proposed approach in detail, followed by experimental results and discussion in 3 and 4, after which we conclude the paper with prospective future work and directions.
2 Materials and Methods
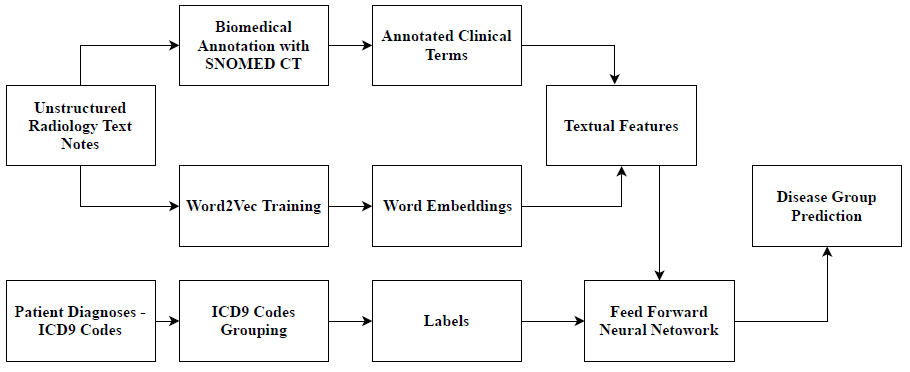
The overall workflow of the proposed approach for disease group prediction is as depicted in Figure 1. Radiology reports in unstructured text format from the open and standard MIMIC-III [11] dataset were used for this study. MIMIC-III contains data about 63,000 ICU admissions of around 46,000 patients admitted in Beth Israel Hospital, New York, USA between 2001 and 2012.
From the 'NOTEEVENTS' table, only the Radiology notes were extracted for this study. Overall, 1,94,744 radiology text reports generated during 45,512 admissions of 36,447 patients were included for the study. Often, a patient may be diagnosed with multiple diseases in the same admission, hence, it is necessary for the prediction to be a multi-label prediction task. Therefore, for each radiology report, all disease groups were considered as labels and given binary values - 0 (if the disease was not present) and 1 (if the disease was present).
Table 1 Dataset and Cohort Characteristics
| Feature | Total Records |
|---|---|
| Patients | 36,447 |
| Admissions | 45,512 |
| Radiology Reports | 194,744 |
| Sentences | 539,466 |
| Words | 45,755,992 |
| Average word Length of Report | 235 |
| Unique Diseases | 2,593 |
| Disease Groups | 21 |
2.1 Preprocessing & Textual Feature Modeling
The radiology reports text corpus were first subjected to a basic Natural Language Processing (NLP) pipeline consisting of tokenization and stopping processes. The tokenization process breaks down the clinical text corpus into tokens and the stopping process filters out unimportant words (stop words) from the corpus. The preprocessed tokens corpus is then fed into a SNOMED-CT ontology based annotator to annotate and extract clinical and biological/biomedical terms. SNOMED-CT ontology [18] is an ontology that provides a vocabulary of clinical/biomedical terms and helps extract associated concepts from the preprocessed radiology report corpus. We used the Open BioMedical Annotator [12] for this purpose, after which 4,366 unique clinical/biomedical terms were obtained.
The presence or absence of each extracted clinical/biomedical term, represented as binary values, is considered as a textual feature representation.
The preprocessed corpus is also used to train a Word2Vec [14] word embedding model to extract the word embedding features from the corpus. The skipgram model of Word2Vec was used for training the corpus as this model takes word ordering into consideration and is effective with infrequent words as well. The skipgram Word2Vec model is trained with a dimension size of 500 and initial learning rate of 0.01. The word embeddings were extracted such that each report is represented as 1 × 500 vector. The word embedding features were further concatenated with the extracted clinical/biomedical term features with binary values for each report indicating its presence (1) or absence (0) in the respective reports and the feature matrix was then standardized to values between -1 and 1. These features are used for training the neural network model for disease prediction.
2.2 ICD9 Disease Code Grouping
The ICD9 disease codes of patients' diagnoses were retrieved from the 'DIAGNOSESJCD' table of MIMIC-III dataset and the labels were grouped as per available standards2 and as previously followed by state-of-the-art work [17]. A total of 2,593 unique ICD9 disease codes were accordingly grouped into 21 ICD9 disease groups (as shown in Table 2). As a patient can suffer from multiple diseases, we consider the ICD9 group prediction task as a binary classification of multiple labels. Therefore, 21 different labels (disease groups) were considered with possible binary values: 0 (for absence of the disease) and 1 (for presence of the disease). The radiology reports of the selected cohort did not have any case of external injury (E-codes), hence, for the 194744 × 4966 feature matrix, 20 ICD9 disease groups were considered as labels to train the neural network model, which is described in Section 2.3.
Table 2 ICD9 Disease Grouping - Dataset Statistics
| ICD9 Group (Label) |
ICD9 Code Range |
Description | Occurrences (MIMIC-III) |
Occurrences (Study sample) |
|---|---|---|---|---|
| 1 | 001 - 139 | Infectious and Parasitic Diseases | 13,686 | 7,289 |
| 2 | 140 - 239 | Neoplasms | 9,457 | 68,734 |
| 3 | 240 - 279 | Endocrine, Nutritional, Metabolic, Immunity | 44,671 | 34,668 |
| 4 | 280 - 289 | Blood and Blood-Forming Organs | 16,345 | 128,357 |
| 5 | 290 - 319 | Mental Disorders | 15,053 | 80,337 |
| 6 | 320 - 389 | Nervous System and Sense Organs | 13,327 | 62,963 |
| 7 | 390 - 459 | Circulatory System | 96,749 | 65,149 |
| 8 | 460 - 519 | Respiratory System | 31,984 | 152,159 |
| 9 | 520 - 579 | Digestive System | 25,206 | 107,656 |
| 10 | 580 - 629 | Genitourinary System | 21,884 | 84,346 |
| 11 | 630 - 677 | Pregnancy, Childbirth, and the Puerperium | 524 | 89,305 |
| 12 | 680 - 709 | Skin and Subcutaneous Tissue | 5,791 | 601 |
| 13 | 710 - 739 | Musculoskeletal System and Connective Tissue | 7,951 | 29,046 |
| 14 | 740 - 759 | Congenital Anomalies | 3,429 | 39,703 |
| 15 | 760 - 779 | Conditions Originating in the Perinatal Period | 19,605 | 10,115 |
| 16 | 780 - 789 | Symptoms | 13,965 | 71,784 |
| 17 | 790 - 796 | Nonspecific Abnormal Findings | 3,034 | 20,803 |
| 18 | 797 - 799 | Ill-defined and Unknown Causes of Morbidity and Mortality | 947 | 6,664 |
| 19 | 800 - 999 | Injury and Poisoning | 30,573 | 108,867 |
| 20 | V Codes | Supplementary Factors | 50,318 | 100,310 |
| 21 | E Codes | External Causes of Injury | 14,003 | 0 |
2.3 Disease Prediction Model
The feature matrix with both word embedding features and ontologically extracted term-presence features, along with ICD9 group labels are next used for training a neural network based prediction model. A Feed Forward Neural Network (FFNN) architecture was used to build the prediction model, which is depicted in Figure 2.
The input layer consists of 2048 neurons with input dimension as 4966 (number of input features); 4 hidden layers with 1024, 512, 256 and 128 neurons respectively and finally an output layer with 20 neurons, each representing an ICD-9 disease group. To prevent overfitting, two dropout layers, with a dropout rate of 20% was also added to the FFNN model (see Figure 2).
As this is a binary classification for multiple labels, the loss function used for the FFNN was binary cross entropy. Stochastic Gradient Descent (SGD) was used as the optimizer and a learning rate of 0.01 was used. The tanh activation function was used as the input and hidden layer activation functions as the feature matrix values are standardized to the range -1 and 1. The major hyperparameters for the FFNN model -the optimizer, learning rate of the optimizer and the activation function, were tuned empirically over several experiments using the GridSearchCV function in Python sklearn library. Finally, the output layer activation function was a sigmoid function, again as the classification was binary for each of the 20 labels. Training was performed for 50 epochs and then the model was applied to the validation set to predict disease groups after which the results were observed and analyzed.
3 Experimental Results
To evaluate the performance of the proposed model, standard metrics to measure machine learning models were considered - accuracy, precision, recall, F-score, Area Under Receiver Operating Characteristic curve (AUROC), Area Under Precision Recall Curve (AUPRC) and Matthew's Correlation Coefficient (MCC). We performed the evaluation of these metrics on a sample-wise basis, i.e., the predicted and actual ICD9 disease groups were compared and analyzed for each radiology report. It can be observed from the Table 3 that, the proposed model achieved promising results: AUPRC of 0.74 and AUROC of 0.84. The accuracy of 0.77 and and precision of 0.80 also indicate effective prediction performance of the proposed approach.
Table 3 Experimental Results
| Parameter | Proposed Approach | Purushotham et al. [17] |
|---|---|---|
| Total admissions | 45,512 | 38,425 |
| Type of Data | Unstructured Text | Structured data |
| AUROC | 0.84 ± 0.01 | 0.77 ± 0.01 |
| AUPRC | 0.74 ± 0.01 | 0.60 ± 0.02 |
| Accuracy | 0.77 | * |
| Precision | 0.80 | * |
| Recall | 0.77 | * |
| F-Score | 0.77 | * |
| MCC | 0.50 | * |
* Metric not reported in the study
We also compared the performance of the proposed approach against an existing ICD9 disease group prediction model, a Multimodal Deep Learning (MMDL) architecture put forward by Purushotham et al. [17], which is built on structured patient data. As the number of records and features under consideration for both the studies are different, it is to be noted that this is a metric based comparison. During validation experiments, it was observed that the proposed approach significantly outperformed against the state-of-the-art method by 23% considering the AUPRC metric and 9% in terms of AUROC. To encourage other comparative studies, certain additional experiments were made.
We also provide the Recall & F-Score performance as well as the MCC values of the proposed model over our easily reproducible patient cohort dataset. The model showed good results in these experiments, achieving a recall of 0.77, F-score of 0.77 and MCC value of 0.50. It is to be noted that our method performed better than the state-of-the-art [17], despite being built on a significantly larger number of patient admission data than the state-of-the-art approach (see Table 3). Further, we achieve this performance using only textual features and we did not make use of structured patient data or processed information from any kind of structured data to model the radiology reports of patients. Thus, there is an added advantage that the conversion from unstructured text data to a structured representation can be ignored, thereby achieving huge savings in person hours, cost and other resources.
4 Observations and Discussion
From our experiments, we observed a very high requirement and potential of developing prediction based CDSSs using unstructured text reports rather than the usage of structured patient data and EHRs. The proposed text feature modeling was effectively able to capture the rich and latent clinical information available in unstructured radiology reports, and the neural network model used these features to effectively learn disease characteristics for prediction. The Word2Vec model generated word embedding features and the extracted terms using the Open Biomedical Annotator and SNOMED-CT ontology further enhanced the semantics of the textual features thereby enabling the FFNN to generalize better and learn the feature representation well resulting in effective prediction performance of the proposed approach.
The high values of metrics AUPRC of 0.74 and AUROC of 0.84 in comparison to the state-of-the-art model's (built on structured data) AUPRC of 0.60 and AUROC of 0.77 respectively, is an indication that the unstructured text clinical notes (radiology reports in this case) contain abundant patient-specific information that can be used for predictive analytics applications and that the conversion process from unstructured patient text reports to structured data can be eliminated thereby saving huge person hours, cost and other resources. Moreover, the proposed approach also eliminates any dependency on structured EHRs, thus making it suitable for deployment in developing countries.
Few other insights into the challenges also came to light during our experiments. We found that the initial data preparation approach used for this study could be improved, as it resulted in some conflicting cases during training. In the MIMIC-III dataset, the radiology reports do not have a direct link to ICD9 disease code and to overcome this, we designed a data preparation approach for extracting ICD9 codes from the DIAGNOSES_ICD table and then assign them to all patients with the same SUBJECT_ID and HADM_ID in the radiology notes corpus.
A negative effect of this approach was that, in some cases, the ICD9 disease codes/groups were assigned to radiology text reports were not related to that particular disease. This could have affected the model's accuracy, due to assignment of conflicting labels to textual features of radiology notes. Nevertheless, the model achieved promising results, as is evident in the values of metrics like precision (0.80), accuracy (0.77), F-score (0.77), recall (0.77) and MCC (0.50). The AUROC (0.84) and AUPRC (0.74) values also show that a disease prediction model built on unstructured radiology text reports can perform well as a real-world CDSS application for hospitals and caregivers.
5 Conclusion and Future Work
A neural network based model for predicting ICD9 disease groups from unstructured radiology text reports was presented in this paper. The approach is built on a feature set modeled using Word2Vec to generate word embedding features and also SNOMED-CT ontology based annotator to extract clinical/biomedical terms and concepts whose presence or absence are considered as features. The ICD9 disease codes were categorized into 21 standard groups and then used to train a binary classifier for multi-label prediction.
A FFNN architecture was used to train the classifier and the prediction model was validated and benchmarked against state-of-the-art ICD9 disease group prediction model. The experiments highlighted the promising results achieved by the proposed model, outperforming the state-of-the-art model (built on structured patient data) by 23% in terms of AUPRC and 9% in terms of AUROC. This indicates that the proposed approach can be considered as a viable alternative, as it eliminates the dependency on structured clinical data, thereby ensuring that hospitals in developing countries with low EHR adoption rate can also utilize effective CDSS in their functioning.
As part of future work, we plan to address the issues observed in the designed data preparation strategy, and enhance it by sorting out the disease group assignment problems. We also intend to explore other feature engineering techniques to further optimize topic and feature modeling representations for their effect on disease prediction.











 nueva página del texto (beta)
nueva página del texto (beta)