1 Introduction
Modern medical studies increasingly use nonstan-dard sources of information to obtain new data related to medical conditions, efficiency of drugs, their adverse effects, interactions between different drugs, and so on. One such source of information can be provided by the drug users themselves, in the form of free-text web reviews, social media posts, and other user-generated texts. These sources have been successfully used, for instance, to monitor adverse drug reactions (ADRs), making it possible to detect rare and underestimated ADRs through the users complaining about their health on social networks or specialized forums [36].
However, it may be important for the medical field to learn more than just the existence of an adverse reaction from a text review. Drugs may exhibit different behavior on people with different age, gender, or other parameters that will often be unknown for a text scraped from an Internet forum. Hence, the problem arises to mine demographic information from free-text medical reviews.
In this work, we make the first steps in the direction of extracting demographic information from user-generated texts related to medical subjects. We have collected databases of medical reviews from health-related Web sites with user-generated content, namely WebMD and AskaPatient, and have trained models to predict the age and gender of users who wrote these reviews. We propose a classification approach based on a classical classifier (we compare SVM and Maximum Entropy classifier, i.e., logistic regression) which is augmented by sets of features based on recently developed novel approaches to text mining: topic models, including the Partially Labeled Topic Model, and features based on word embeddings. We show that the resulting classifier performs significantly better than the baseline.
This work is a significantly extended journal version of the paper [40]; compared to the conference version, we have changed the approach to baseline classifiers, making them into feature-rich classifiers with topics and word embeddings as features. We have also significantly extended the set of said features, adding new domain-specific information to aid the classifiers. Therefore, the experimental part of this work is new compared to [40], and the results have been substantially improved.
The paper is organized as follows. In Section 2, we survey related work on mining drug-related information from social media and other user-generated texts. Section 3.1 defines models for information extraction from text that we compare in this work: we present the features and briefly introduce topic models with user attributes and distributed word representations. We present experimental results in Section 4 and conclude with Section 5.
2 Related Work
The use of social media for medical and pharma-cological data mining has been on the uprising since early 2010s; the term “pharmacovigilance” has been coined for automated monitoring of social media for potentially adverse drug effects and interactions; see also media articles about these effects [14, 37]. One of the first works on this subject [13] analyzed user posts regarding six drugs from a health-related social network. A comprehensive review of text mining techniques as applied to drug reaction detection can be found in [9]. We also note a Social Media Mining Shared Task Workshop (organized as part of the Pacific Symp. on Biocomputing 2016) devoted to mining pharmacological and medical information from social media, with a competition based on a published dataset [35].
In [6], authors identify ADRs from texts on health-related online forums. They used dictionary-based drug detection, extracting symp-toms with a combination of dictionary-based and pattern-based methods. A lift measure (also known as pointwise mutual information) was computed to evaluate the likelihood of drug-ADR relation and chi-square test was used to evaluate the statistical significance of the lift measure. Several case studies of drugs showed that some ADRs were reported prior to FDA approval. One limitation of this work is the number of annotated examples in test data: less than 500 ADRs for evaluation.
In [32], existing machine learning dictionary-based approaches were used to identify disease names from user reviews about top 180 most frequently searched medications on the forum WebMD, using a rule-based system to extract beneficial effects of the drug. In order to identify candidates for drug repurposing, authors removed known drug indications and manually reviewed the comments without FDA reports. The main limitation of this work is the lack of an annotated corpus to evaluate the proposed method. The work [42] shows an experiment for ten drugs and five ADRs to examine associations between them on texts from online healthcare communities using association mining techniques. The FDA alerts served as a gold standard to evaluate the associations discovered between drugs and ADRs. We also note a series of works specifically on Spanish language social media [15,36].
Usually, pharmacovigilance studies employ simple classifiers to extract information on drug effects or interactions. For example, to mine drug-related information from a stream of Twitter data, a recent work [24] uses a cascade of simple input filters followed by an SVM classifier, reporting good discovery results, while [44] proposes a weighted average ensemble of four classifiers: one based on a handmade lexicon, two on n-grams, and one on word embeddings.
On the other hand, drug testing and discovery of drug effects and interactions requires one to know demographic information about a user since drug effects can differ significantly depending on the user. This leads to the need to mine demographic information about the authors together with the user-generated texts themselves. When such information is provided, e.g., when the texts are collected from facebook users with explicitly known age and gender, there is no problem. However, in many situations user reviews for drugs and medical services are found anonymously on review web sites such as WebMD or AskaPatient; often demographic information can be known for a minority of users but not all. Hence, the problem arises to predict user demography based on the texts of user reviews.
In natural language processing, predicting demographic features based on free text falls into a large classical field of authorship analysis, attribution and author verification studies [12, 45]; we refer to surveys [3, 38, 39] for details and references. Numerous works on the topic have been published based on the results of the shared Author Profiling Tasks at digital text forensics events by PAN initiative [2, 5, 7, 27–30]. However, authorship analysis seldom extends to medical issues: for example, the work [23] attempts to screen Twitter users for depression based on their tweets, but to the best of our knowledge, previous work has not attempted to automatically mine demographic information unless it was provided explicitly. In this work, we begin to fill this gap, providing first results on automated predictions of demographic based specifically on medical reviews.
3 Classification Methods
3.1 Models
In this section, we describe two different ap-proaches for demographic prediction applied to a collection of user comments about medications. First, we describe our feature-rich machine learning classifiers. Second, we describe neural networks that rely on word representations learned from unannotated reviews.
3.2 Basic Classifiers and their Features
We formulate the prediction of user attributes as a classification problem. In order to perform the classification, we apply two supervised approaches with a set of hand-crafted features:
(1) support vector machine (SVM);
(2) logistic regression, also called the Maximum Entropy classifier (MaxEnt).
These approaches have been known to achieve the best results in various classification tasks, including sentiment and subjectivity classification [11, 41], ADR classification [34], and demographic prediction [22, 31]. Our classifiers leverages a variety of surface-form, semantic, cluster-based, distributed and lexicon features described below.
The entire set of features used in our classifiers consists of the following subsets:
— Word ngrams (NGR): occurrence of contiguous sequences of 1, 2, and 3 tokens; the maximum number of features are 25,000;
— Drug classification groups (ATC): drug names are classified in groups at five different levels using the DrugBank database and the ATC classification system;
— Automatically generated lexicons (PMI): for each token occurring in a text and present in our automatic lexicon, we use its score to compute the number of tokens with score(w) > 0 and sum of these scores, the number of tokens with score(w) < 0 and sum of these scores, the total score, and maximal and minimum scores; all scores and sums are averaged for each review;
— Sentiment lexicons (SENT): for each of the sentiment lexicons (Bing Lius Lexicon and MPQA Subjectivity Lexicon), we compute the following two features: average sum of positive scores for the tokens and average sum of negative scores for the tokens;
— ADR lexicon (ADR): presence/absence of ADR mentions using the lexicon;
— Clusters (CL): presence/absence of tokens from each of the 150 clusters;
— Topics (TPC): presence/absence of tokens from each of 150 topics;
— Word embeddings (EMB): the real-valued vector of each word as described in Section 3.4.
In the remainder of this subsection, we define each of these items in detail.
ATC classification. In the Anatomical Ther-apeutic Chemical (ATC) classification system, biomedical and chemical entities are divided into different groups according to the organ on which they act and their therapeutic, pharmacological, and chemical properties. Using the DrugBank database, we find the presence of a drug in each class up to 5 levels. For example, Prozac (Fluox-etine) is associated with the ATC code N06AB03 and classified into this code and the following codes from higher levels: ’elective serotonin reup-take inhibitors’ (N06AB), ’antidepressants’ (N06A), ’psychoanaleptics’ (N06), ’nervous system’ (N). We use these features to incorporate domain-specific medical knowledge into the classification process.
Automatically generated lexicon. The key idea of this automatically generated lexicon is to take advantage of a large corpus of weakly labeled texts, where authors assign several predefined labels to each text. Following state-of-art approaches for sentiment analysis [11], we automatically generated a lexicon based on the score for each token (w) (with frequency greater or equal than 10) in the Health dataset:
where
is the pointwise mutual information, cat denotes all texts associated with the particular category, oth denotes all texts in other categories, and p(w, pt) are probabilities of w occurring in the texts labeled with a particular category. As categories we separately use age and gender attributes.
Sentiment lexicons. We used Bing Lius Lexicon1 and the MPQA Subjectivity Lexicon2. We assign the score of +1 for positive entries and the score of -1 for negative entries from the Bing Lius Lexicon. For the MPQA Subjectivity Lexicon, we assign scores +0.5/-0.5 and +1/-1 for weak and strong associations respectively.
ADR lexicon. We assume that patients experience different adverse drug reactions that may depend on age and gender. In order to use medical information specific to demographic groups, we develop the exact lookup based on ADR lexicon from the paper [34]. The lexicon contains 16,183 ADRs from several resources: the COSTART vocabulary created by the FDA for post-market surveillance of ADRs, the SIDER side effect resource, and the Canada Drug Adverse Reaction Database, SIDER II and the Consumer Health Vocabulary.
Cluster-based features. Clusters reduce the sparsity of the token space as an alternative rep-resentation of text. We use the Brown algorithm, i.e., a hierarchical clustering algorithm [4]. The algorithm partitioned the words into a set of 150 clusters, and we add features corresponding to the presence or absence of specific clusters in the review.
Next, we discuss the last two classes of features that come from topic models and word embeddings respectively.
3.3 Topic Models
For topic-based features, we employ the latent Dirichlet allocation (LDA) model, a classical topic model. We assume that a corpus of D documents contains T topics expressed by W different words. Each document d ∈ D is modeled as a discrete distribution θ(d) on the set of topics: p(zw = t) = θtd, where z is a discrete variable that defines the topic of each word w ∈ d. Each topic, in turn, corresponds to a multinomial distribution on words: p(w | zj = t) = φwt (here w denotes words in the vocabulary and j denotes individual instances of these words). The probabilistic graphical model of basic LDA is shown on Fig. 1a. The model introduces Dirichlet priors with parameters α for topic vectors θ, θ ∼ Dir(α), and β for word distributions φ, φ ∼ Dir(β) (we assume the Dirichlet priors are symmetric, as they usually are). A document is generated word by word: for each word, first sample its topic index t from θd, t ∼Mult(θd), then sample the word w from φt, w ∼Mult(φt). We denote by nw,t,d the number of words w generated with topic t in document d; partial sums over such variables are denoted by asterisks, e.g., n∗,t,d = Σw nw,t,d is the number of all words generated with topic t in document d, nw,∗,∗ = Σt,d nw,t,d is the total number of times word w occurs in the corpus and so on; we denote by ¬j a partial sum over “all instances except j”, e.g., 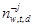 is the number of times word w was generated by topic t in document d except position j (which may or may not contain w). In the basic LDA model, inference proceeds with collapsed Gibbs sampling, where θ and φ variables are integrated out, and zj are iteratively resampled as follows:
is the number of times word w was generated by topic t in document d except position j (which may or may not contain w). In the basic LDA model, inference proceeds with collapsed Gibbs sampling, where θ and φ variables are integrated out, and zj are iteratively resampled as follows:
where z−j denotes the set of all z values except zj. Samples are then used to estimate model variables:
We also experimented with Partially Labeled Topic Model (PLDA) [26]. PLDA incorporates user meta-data tags (e.g., location, gender, or age) together with topics. In this model, each document is assigned with an observed tag or a combinations of tags, topics are generated conditioned on the document’s tags, and words are conditioned on the latent topics and tags. The probabilistic graphical model of PLDA is shown on Fig. 1b. The Gibbs sampling step proceeds as
An important characteristic feature of topic models is that they can be mined for qualitative results that are easy to interpret and can validate their performance. For example, Table 1 shows topics discovered by the PLDA model based on a unigram representation of reviews related to each gender; note that the distinction between ”male” and ”female” topics does indeed reflect common medical knowledge.
3.4 Distributed Word Representations
The other class of models in our study is very different in nature from topic models. We compare results produced by topic models with classifi-cation models based on word2vec embeddings processed by recurrent and convolutional neural networks (RNNs and CNNs).
Recent advances in distributed word represen-tations have made it into a method of choice for modern natural language processing [8]. Distributed word representations are models that map each word occurring in the dictionary to a Euclidean space, attempting to capture semantic relationships between the words as geometric relationships in the Euclidean space. In a classical word embedding model, one first constructs a vocabulary with one-hot representations of individual words, where each word corresponds to its own dimension, and then trains representations for individual words starting from there, basically as a dimensionality reduction problem. For this purpose, researchers have usually employed a model with one hidden layer that attempts to predict the next word based on a window of several preceding words. Then representations learned at the hidden layer are taken to be the word’s features.
The word2vec embeddings come in two flavors, both introduced in [16]: Continuous Bag-of-Words (CBOW) and skip-gram. During its learning, a CBOW model is trying to reconstruct the words from their contexts with a network whose architecture is shown on Fig. 2a; the training process for this model proceeds as follows:
(1) each of the inputs of this network is a one-hot encoded vector of size |V|, where V is the vocabulary;
(2) when computing the output of the hidden layer, we take an average of all input vectors; the hidden layer is basically a matrix of vector embeddings of words, so the nth row represents an embedding of the nth word in the vocabulary;
(3) the output layer represents a score uj for each word in the vocabulary; to obtain the posterior, which is a multinomial distribution, we then use the softmax
so the loss function is
The skip-gram model operates inversely, predicting the context from the word, which can be seen from its network architecture shown on Figure 2b. Here the target is an input word, and the output layer, in turn, now represents C multinomial disctibutions
with the loss computed as
The idea of word embeddings has been applied back to language modeling in [17, 18, 21], and starting from the works of Mikolov et al. [16, 19], word representations have been used for numerous NLP problems, including text classification, extraction of sentiment lexicons, part-of-speech tagging, syntactic parsing, and others.
Word embedding models represent each word using a single real-valued vector. Such represen-tation groups together words that are semantically and syntactically similar [20]. We used word2vec from Gensim library to train embeddings on the Health Dataset. We applied Continuous Bag of Words model with the following parameters: vector size of 200, the length of local context of 10, negative sampling of 5, vocabulary cutoff of 10. Below, we refer to our pre-trained vectors as HealthVec. We also experimented with another published word vector PubMedVec (2,351,706 terms) trained on biomedical literature indexed in PubMed [25]. PubMed comprises more than 26 million citations for biomedical literature from bibliographic database MEDLINE, journal articles, life science journals, and online books.
3.5 Neural Network Classifiers
In this work, we compare two modern approaches to natural language processing with neural networks: traditional recurrent architectures, specifically LSTM-based recurrent networks, and convolutional neural networks (CNN). In the recurrent part, we use an architecture with multiple LSTM layers, where higher layers use the sequence of outputs from the previous layer of LSTMs, and on the top level the LSTM outputs are combined into the final layer which does the actual prediction.
While CNNs have been most successfully used for image processing, recent applications of CNNs to natural language processing also produce state of the art results. In an NLP task, convolutional layers are still interleaved with subsampling max-pooling layers, but this time the convolutions are one-dimensional rather than two- or three-dimensional as in images and video. Here, we use a convolutional model similar to the one recently presented in [10] for semantic sentence classification; this model has the following characteristic features:
— it is not as deep as computer vision models and involves only one convolutional layer with max-over-time pooling and a softmax output;
— regularization is achieved through dropout; the authors report a consistent and significant improvement in accuracy with dropout across all experiments;
— the model is trained on prepared word2vec word embeddings and does not attempt to tune word representations for better results;
— still, the authors report better results on such tasks as sentiment analysis and sentence classification than baseline techniques that include recursive autoencoders and recursive neural networks with parse trees.
4 Evaluation
4.1 Datasets
For experimental evaluation, we have crawled health-related reviews from two health hotel review sites: (i) WebMD3 and (ii) AskaPatient4.
WebMD is a health information services website that aims to provide objective, trustworthy, and valuable health information. We have crawled 217,485 reviews from authors tagged as “Patient”. Each review contains the following fields: 1) date when the review was written, 2) condition for taking treatment, 3) free-text review given for the effects caused due to the use of the drug, 4) user attributes such as gender and age. Gender tags are “Male” or “Female”, and predefined age tags in the dataset are “19–24”, “25–34”, “35-44”, “45-54”, “55-64”, “65-74”, or “75 or over”. In this study, we combine some of the age tags and divide user attributes into three major age groups: “19-34”, “35-64”, and “65 and over”.
AskaPatient5 website aims to empower patients by allowing them to share and compare their medical experiences. We have crawled 113,093 reviews. Since users often confuse two free-text fields about a drug, we have concatenated the “side effects” and “comments” fields, treating the result as a full review. Similar to WebMD, reviews from AskaPatient contain textual information, reason for taking treatment and user attributes (without predefined list of age groups).
In contrast with our previous work, we split our corpora into training and testing parts further referred as WebMD and AskaPatient (used by the ML and DL algorithms) and a free-text corpus of in-domain texts called the Health dataset (used to compute PMI, topics, and word representations). In order to create robust methods and exclude drugs with highly imbalanced genders (e.g., birth control pills), we use reviews associated with 5 most commented conditions for training/testing. For WebMD, review authors select a condition from a predefined list for every drug. For AskaPatient, the “reason” is a free-text field.
Table 2 summarizes the statistics of both datasets used in our study. The WebMD dataset contains 20,693 reviews with the age group “35-64”, 7,410 reviews with the age group “19-34”, and 7,519 reviews with the age group “65 and over”. The total numbers of tokens in the WebMD and AskaPatient datasets are 2,818,429 and 1,051,969, respectively. The total numbers of unique tokens in the WebMD and AskaPatient datasets are 33,411 and 18,825, respectively.
4.2 Model Parameters
In order to get local features from a review with CNNs we have used multiple filters of different lengths [10]. We separated out 10% of the training dataset to form the validation set which was used to evaluate different model parameters. We used a sliding max-pooling window of size 2 to get features through filters. Pooled features are then fed to a fully connected feed-forward neural network (with dimension 100) which uses rectified linear units as output activations. Then we apply a softmax classifier with the number of outputs equal to the number of classes. We applied dropout rate of 0.5 to the fully connected layer and trained the network for 20 epochs; on the other hand, we did not apply dropout after the embedding layer since in our experiments this led to lower results achieved by CNNs on the validation set. The width of the convolution filters is set to 3, each with 100 filters. Additionally, we employ early stopping after two epochs with no improvement on the validation set. Embedding layers are not trainable for all networks. We set mini-batch size to 256 and 128 for the WebMD and AskaPatient datasets, respectively.
In our experiments with recurrent neural net-works, we used a standard GRU or LSTM architecture on top of the embedding layer that implemented pre-trained word embeddings. Similar to [1], the resulting sequence of vectors serves as the input to the network. We experimented with shallow GRU/LSTM, two-layer GRU, and used 100 units on each layer with the Adam optimizer and rectified linear units as output activation. Similar to CNN, GRU layer is then fed to a fully connected feed-forward neural network (with dimension 100). Other parameters are adopted from CNN settings.
We tested and compared the following vectors:
The general statistics are presented in Table 3. We also observed better classification results after normalizing each vector by dividing it by its 2-norm.
For SVM and MaxEnt classifiers, we used LinearSVC and LogisticRegression with default parameters from the NLTK library7. We used Liang’s implementation of the Brown hierarchical word clustering algorithm8.
We used the Mallet9 library to generate topics. The number of sampling iterations was set to 1000. We used default hyperparameters, took top 20 words for each topic, and evaluated 50, 100, and 150 topics on the validation set. The best results were achieved with 150 topics. We also implemented PLDA for comparison, adopting its parameters from LDA. For further evaluation, we selected topics from LDA rather than PLDA since they produced better results on validation data.
4.3 Results
In this section, we describe our experiments with feature-rich classifiers and deep learning models. We performed pre-processing by lower-casing all words. We performed 5-fold cross-validation and computed precision (P), recall (R), and F1-measure (F1), showing the macro-averaged results in Table 4 (gender prediction) and Table 5 (age prediction). The tables also show the best results for each model type in every column highlighted in bold.
The main result, which might look surprising at first, is that standard classifiers, when enriched with a large number of various features, outperform even the best neural network approaches that we have been able to train. Specifically, CNNs and RNNs are able to achieve better precision than SVM and MaxEnt but lose significantly in recall and therefore in the aggregate F1 measure. Moreover, Tables 4 and 5 show the variances of the F1-measure in our cross-validation results, indicating that the advantage of SVM and MaxEnt in F1-measure is statistically significant.
This seemingly unexpected result is, in our opinion, due to two main reasons. First, we are free to augment standard classifiers with any features we want, thus using a wide variety of external information that is unavailable to the neural networks, which have to rely on text only. It is unclear how to introduce all of the features that we used for SVM and MaxEnt into the neural networks, and it would require a separate complex study, both theoretical and practical, to incorporate these features.
Second, the dataset size in this case is probably not large enough for the neural networks to shine. Since we used suitable regularization we did not experience strong overfitting in the neural networks, but general rules of thumb suggest that our supervised datasets are too small for the expressive power of complex neural networks to have significant effect.
Thus, our results suggest that while neural network approaches often define the state of the art in modern natural language processing, in problems where rich additional information can be made available, especially in domain-specific problems with well defined domains (such as medicine in this case), classical machine learning approaches can still be very useful and can still be successfully used in practical settings.
Secondary results include two conclusions from Tables 4 and 5. First, while adding more features is usually obviously beneficial, this did not hold for ATC features in our experiments: they helped much less than others and even deteriorated the results. This is probably due to the fact that a relatively small dataset size combined with high dimension of ATC features led to overfitting. Second, note that the best results with neural networks are usually obtained in variations where the word embeddings are also trainable. Regardless of the dataset size (which would be much too small to train embeddings properly), in our experience making embeddings trainable (i.e., slightly fitting them in the end-to-end supervised network starting from unsupervised vectors) appears to be beneficial almost always and should be adopted in most settings.
We have also performed qualitative analysis of our results. In particular, we have extracted and analysed the most representative n-grams for various conditions. Tables 6 and 7 present the most representative features (excluding numeric features) for one gender over another and for a certain age group over other age classes, respectively. For this experiments, we used the MaxEnt classifier trained on the set of 2- and 3-grams extracted from the review texts. The tables indicate that key terms change with age or gender, reflecting quite natural progressions that match well with medical and commonsense intuition. Hence, our classifiers can also be used to mine qualitative information from a dataset of medical reviews, perhaps uncovering new common conditions or important factors in a certain user group.
Table 6 Representative MaxEnt features for male and female patients with different conditions (WebMD dataset)

5 Conclusion
In this work, we have presented the first results on the practically important problem of automatically learning demographic user features from his or her reviews concerning medical products or services.
We have compared several different models for gender classification and age prediction: baseline classifiers that operate on words and bigrams, feature-rich classifiers with additional information from topic models and word embeddings as well as domain-specific medical information, and convolutional and recurrent neural networks based on word2vec embeddings.
Results of our experiments suggest that in settings with relatively small datasets and available external information classical machine learning techniques can outperform neural network ap-proaches. This is due to both dataset size and the fact that while it is hard to tailor neural networks to a specific form of external information, standard classifiers incorporate such new features trivially. We believe that this sample application shows that there is still a place for domain-specific machine learning solutions, especially for relatively small supervised datasets.











 text new page (beta)
text new page (beta)



















