Introduction
Chloroplast and mitochondrial DNA analysis (chloroplast DNA and mitochondrial DNA, respectively) may reveal information about evolutionary processes by phylogenetic analysis using uni-parental inheritance patterns and conserved gene sequences across generations (Ravi et al., 2008; Jo et al., 2011). It may also describe the composition and organization of the genetic information inside chloroplasts and mitochondria (Kahlau et al., 2006), and provide their genes sequence, order and function (Sugiyama et al., 2005; Kahlau et al., 2006). On this regard, in our lab we have identified two important genetic components that significantly modify pungency expression in Manzano hot pepper (Capsicum pubescens R. y P.). First, Cruz et al. (2007) showed the existence of heterosis in Manzano pungency, and afterwards Sánchez et al. (2010) showed maternal inheritance effects of fruit pungency based entirely on traditional plant breeding techniques. To confirm maternal inheritance effects on the Manzano pungency by a more precise method, DNA had to be extracted from chloroplasts and mitochondria obtained from plenty Manzano pepper fruits. The downstream molecular analysis required the isolation or enrichment of intact organelles in a reliable, accurate and repeatable way.
There are studies related to chloroplast and mitochondrial DNA isolation and extraction (Douce et al., 1987; Hájek et al., 2004; Jansen et al., 2005; Kahlau et al., 2006; Lang et al., 2011; Millar et al., 2001; Mourad and Polacco, 1988; Rahman and Huber, 1996; Rowan and Bendich, 2011; Salvi et al., 2008; Triboush et al., 1998). The methods mentioned above prioritized structural integrity of isolated organelles and low levels of contaminants to ensure high-quality DNA. Some strategies focused on intensity and duration of the homogenization (Hájek et al., 2004), as well as on usage of quality reagents for extraction, washing, purification and lysis. Appropriate buffers keep organelle integrity by maintaining adequate osmotic pressure and pH when cells are broken (Millar et al., 2007). However, the reference protocols recommend a starting sample amount from tens of grams to kilograms of fresh plant tissue. For example, Rahman and Huber (1996) used 100 g of fruit pericarp homogenized in 150 mL of extraction buffer, Kahlau et al. (2006) used 50 g of leaves in 2 L, Shi et al. (2012) used 20 g of leaves in 400 mL, and Vieira et al. (2014) used 25 g of leaves in 400 mL of buffer. These sample amounts could be difficult to obtain, and some protocols address this concern without affecting DNA yield or quality, through a series of strategies. Mourad and Polacco (1989) used 10 g of fresh leaves in 10 volumes of extraction buffer, and O’Hara and Capwell (1993) and Triboush et al. (1998) used 5 g of tissue in 20-25 mL of buffer.
Most methods describing organelle isolation use density gradient centrifugation. This type of purification requires centrifugation after layering the cell homogenate onto a density gradient created by viscous sucrose, percoll or cesium chloride. Proper separation requires ultra-centrifugation to obtain a concentrated organelle fraction (Millar et al., 2007). Recommended gradients have shortcomings, for example,sucrose might enter the mitochondrial matrix when a sucrose gradient is employed, thus hindering proteomics analysis (Nikaido and Rosenberg, 1983; Benz, 1994); and a percoll gradient is occasionally difficult to remove (Beavis and Vercesi, 1992; Yang and Mulligan, 1996).
The most efficient protocols consume high buffer quantities, which significantly increase costs when hundreds of samples are processed, such as in genotyping studies. Our research relies on a miniprep that reduces costs and produces intact chloroplasts and mitochondria destined to DNA extraction in large-scale studies. The proposed miniprep allows processing numerous samples, in our case Manzano chili samples, while reducing volumes of reagents and using rather common lab equipment.
Materials and methods
Plant material
Samples for this research were taken from inbred lines of Manzano pepper derived from previous breeding and selection efforts. Originally, commercial varieties Zongolica and Puebla were crossed, and an F2 population derived by selfing. From this F2 population, two individuals contrasting in spiciness were selected. From both selected individuals, an inbred line was created by 5 selfing cycles. The resulting lines were then crossed both ways (i.e. L4×L6 and L6×L4), and the F1 progeny was sampled. The above description depicts a common genotyping experiment that requires analisys of a high number of individuals. All Manzano pepper plants were grown in a greenhouse, in accordance with the management techniques recommended by Pérez-Grajales and Castro-Brindis (2008) for this plant species. Fruits were sampled at the beginning of the ripening process when the pericarp color changes from green to yellow. Fresh harvested fruits were washed with soap and rinsed with distilled water. A 500 mg pericarp sample was taken and cut in small pieces.
Chloroplast enrichment
The method proposed in this research is based on Kahlau et al. (2006). The tissue was kept in 2 mL plastic tubes on ice. Steel disruptor balls and 1 mL of cold extraction buffer (350 mM sorbitol, 50 mM Tris-HCl pH 8.0, 5 mM EDTA, 15 mM β ME and 0.1 % BSA) were added. Tissue was disrupted using a Tissuelyzer (QIAGENTM, Germany), set at 30 hertz, for 3.5 min. The homogenate was filtered through a nylon net (0.25 mm hole size) into a clean 2 mL test tube; the nylon net and its contents were discarded. The cell debris and nuclei in the filtered homogenate were pelleted by spinning at 500 xg for 5 min at 4 °C. The supernatant was again transferred to a clean 2 mL test tube, and then used for chloroplast enrichment. All samples and reagents were kept cold in an ice bath. The supernatant from the previous step was centrifuged at 2000 xg for 10 min at 4 °C. After centrifugation, the supernatant was removed carefully without touching the chloroplast pellet at the bottom. The pellet was dissolved and washed in 500 (L of cold wash buffer (350 mM sorbitol, 50 mM Tris-HCl pH 8.0, 25 mM EDTA and 0.1 % BSA). The suspension was washed and centrifuged at 2000 xg for 10 min at 4 °C twice, removing the supernatant after each centrifugation. The resulting pellet was suspended in 500 µL of cold wash buffer.
To selectively enrich the suspension for chloroplasts, we employed the density gradient centrifugation method. The gradient was constructed by preparing two molecular-grade sucrose solutions at 1.75 M and 1.08 M. The required sucrose was dissolved in adequate volumes of 50 mM Tris-HCl pH 8.0 and 25 mM EDTA pH 8.0. Seven hundred (700) µL of the 1.75 M sucrose solution were poured into a new 2 mL tube, and then 900 µL of 1.08 M sucrose solution was delicately placed on top of it. Three hundred (300) µL of the chloroplast suspension was gently and slowly placed on top of the sucrose gradient, one drop at a time to avoid mixing. The tube was closed and centrifuged at 7000 xg for 1 hour at 4 °C. After centrifugation, a small, green pellet formed on the sidewall of the test tube; this pellet was carefully collected with a micropipette and placed in a new tube. The pellet was suspended in three volumes of cold dilution buffer (175 mM sorbitol, 50 mM Tris-HCl pH 8.0, 25 mM EDTA). Finally, the tube was centrifuged at 2000 xg for 10 min at 4 °C.
Mitochondria enrichment
Our protocol is based on the method suggested by Rahman and Huber (1996) for mitochondrial DNA isolation. Extra steps were added to improve DNA precipitation and extraction. The tissue was kept in 2 mL plastic tubes on ice. Tissue was disrupted using a Tissuelyzer (QIAGENTM, Germany). Steel disruptor balls and 1 mL of cold extraction buffer (400 mM sucrose, 50 mM Trizma base, 1 mM EDTA, 10 mM KH2PO4, 4 mM cysteine and 1 % BSA; pH 7.6) were added to the tube containing the tissue sample. The Tissuelyzer was set at 30 hertz for 3.5 min, and the homogenate was filtered through nylon net (0.25 mm hole size) into a clean 2 mL test tube; the nylon net and its contents were discarded. Cell debris and nuclei in the homogenate were pelleted by spinning the tube at 500 xg for 5 min at 4 °C. The supernatant was carefully transferred by pipette to a clean 2 mL test tube, and then centrifuged at 2000 xg for 10 min at 4 °C to pellet chloroplasts for removal. This step was repeated twice. After centrifugation, the supernatant was transferred to a new test tube, and mitochondria were pelleted by centrifuging the supernatant at 16000 xg for 10 min at 4 °C. Without disturbing the pellet at the bottom, the supernatant was removed. The mitochondria pellet was then dissolved in 500 µL of cold wash buffer (400 mM mannitol, 10 mM KH2PO4 and 0.5 % BSA, pH 7.2). The suspension was centrifuged at 16 000 xg for 10 min at 4 °C; the wash step was repeated one more time, removing the supernatant after every centrifugation.
Density gradient centrifugation was employed to selectively separate mitochondria from other subcellular organelles. Two sucrose solutions at 0.6 and 1.8 M were prepared by dissolving molecular-grade sucrose into the appropriate volume of 50 mM Tris-HCl pH 8.0 and 10 mM KH2PO4 pH 8.0. The gradient was constructed in a 2 mL test tube by adding 700 µL of 1.8 M sucrose, followed by careful and slow addition of 900 µL of 0.6 M sucrose on top, without mixing. Three hundred (300) µL of mitochondrial suspension were added, one drop at a time to avoid mixing. This tube was centrifuged at 22 000 xg for 1 hour at 4 °C. The mitochondria settled on the tube sidewall as a greenyellow pellet. The mitochondrial fraction was collected with a micropipette and placed in a new test tube. Three volumes of cold dilution buffer (175 mM mannitol, 10 mM KH2PO4 without BSA, pH 7.2) were added, and the test tube was centrifuged at 16 000 xg for 10 min at 4 °C. Steps for both minipreps are briefly illustrated in Figure 1.
DNA extraction
The pellet obtained from chloroplasts or mitochondria enrichment was dissolved in 500 µL of lysis buffer (50 mM TrisHCl pH 8.0, 20 mM EDTA, 2 % N-lauroylsarcosine sodium salt). The mix was kept at room temperature for 15 min to promote organelle rupture. At the end of the incubation time, 1 mL of phenol:chloroform:isoamyl alcohol (25:24:1) solution was added. The tube was shaken in a vortex and then centrifuged at 14 000 xg for 10 min at 4 °C. The supernatant was collected and mixed again with phenol:chloroform:isoamyl alcohol (25:24:1) solution. The DNA was precipitated with 2.5 volumes of cold absolute ethanol and maintained overnight at -20 °C. The next day, the tubes were centrifuged at 14 000 xg for 10 min at 4 °C to pellet DNA. The supernatant was discarded, and the remaining ethanol was evaporated at room temperature for 1 hour. The DNA pellet was dissolved in 30 µL of 0.1 M TE buffer (10 mM Tris, pH 8.0 and 1 mM EDTA), and it was kept at room temperature for 1 hour. DNA integrity was checked on a 0.8 % agarose gel, using 6 µL of each sample per well. The separation conditions were 90 V and 400 mA for 90 min. Gels were then stained with 10 % of ethidium bromide for 15 min and photographed under UV light for documentation. DNA quality was determined in a Nanodrop (Thermo ScientificTM, Mod. 2000, USA) by absorbance.
DNA integrity verification and cross contamination
The extracted DNA from chloroplast and mitochondria fractions was amplified in a MaxygeneTM II thermal cycler (Axygen®, USA). Primers for reference genes were designed based on Genebank information (http://www.ncbi.nlm.nih.gov/genbank/). Three genes were selected for their exclusive location to the nucleus (GenBank accession AY489050.1), chloroplast (GenBank accession AB586585.1) or mitochondria (Genebank accession EU701225.1) (Table 1). Primers were designed according to their coding sequence (CDS) using Primer3 v.4 (http://primer3.ut.ee/) with standard criteria. The PCR reaction mix was setup as follows: 40 ng of DNA, PCR buffer 1x, 2.5 mM of MgCl2, 0.2 mM of DNTP and 20 pM for each primer. The PCR program ran as follows: 95 °C for 3 min, 35 cycles of 95° C for 30 s, 55 °C for 30 s, 72 °C for 40 s, and 72 °C during 10 min. After PCR completion, a 6 μL volume of PCR product per well was separated in a 1.2 % agarose gel in TAE 1x; the running conditions were 90 V and 150 180 mA for 90 min. The gel was stained in 10 % ethidium bromide for 15 min. PCR band size was calculated using LabWorks® software.
Table 1 Primers for reference genes from nucleus, chloroplast and mitochondria.
| Gen | Location | Name | Sequence | ||
| Ubiquitina | Nucleus | CaUBIf | CTGGAAAGCAGCTTGAGGAC | CaUBIr | TGGCCTTAACGTTGTCGATT |
| Rubisco large subunitb | Chloroplast | CaRBCLf | TCACGCTGGTACCGTAGTAGG | CaRBCLr | CTCATTACCTTCCCGAGCAA |
| Cytochrome oxidasec | Mitochondria | SoCOX1f | TACCAGCCATTCTGGAGGAG | SoCOX1r | CTGCCAGTACCGGAAGTGAT |
Primer designed from a Capsicum annuum (GenBank accession AY489050.1), b Capsicum sp. (GenBank accession: AB586585.1), and c Solanum nigrum (GenBank accession EU701225.1) sequences.
Results and discussion
Miniprep step sequences for chloroplast and mitochondria enrichment and their DNA extraction were successfully scaled down to volumes equal or lower than 2 mL. Chloroplast DNA extractions produced average contents of 10.6 µg and an average absorbance ratio A260/A280 of 1.84, while average mitochondrial DNA content was 16.7 µg and A260/ A280 average absorbance ratio of 1.75. These values show higher efficiency for the mitochondrial DNA extraction than for the chloroplast DNA extraction (the average yield difference was 12 µg DNA g-1 fresh tissue) (Table 2). Both DNA samples on agarose gels revealed clear and defined bands without sweeping (Figure 2A).
Table 2 Quantity and quality of chloroplasts and mitochondrial DNA extracted from 500 mg of pericarp from Manzano pepper (Capsicum pubescens R. and P.) fruits at ripening.
| Organelle | Sample | Average DNA quantity (µg) | Average A260/A280 | Average yield (µg DNA g-1 fresh tissue) |
| P1 | 9.09±0.02 | 1.89±0.02 | ||
| Chloroplast | P2 | 13.32±0.04 | 1.73±0.04 | 21.0±4.23 |
| H1 | 11.24±0.09 | 1.96±0.09 | ||
| H2 | 8.78±0.04 | 1.79±0.04 | ||
| P1 | 15.36±0.09 | 1.85±0.09 | ||
| P2 | 14.72±0.12 | 1.72±0.12 | ||
| Mitochondria | H1 | 15.19±0.16 | 1.76±0.14 | 33.4±6.41 |
| H2 | 21.48±0.06 | 1.69±0.06 |
N=3; ± standard deviations; P1: inbred line 4; P2: inbred line 6; H1: hybrid 4×6; H2: hybrid 6×4.
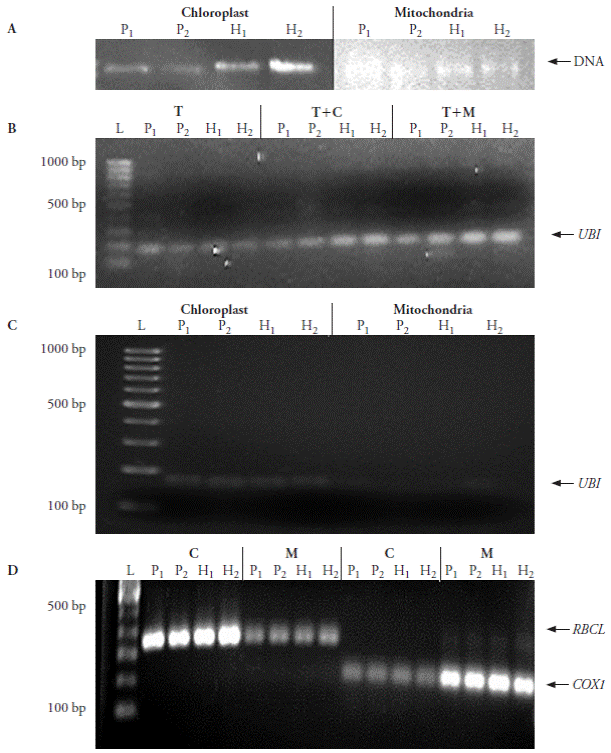
Figure 2 Integrity and purity check for DNA extracted from chloroplast and mitochondria isolated using the proposed method via PCR amplification of reference genes present only in nucleus, chloroplast and mitochondria. A) DNA integrity test. B) Nuclear DNA contamination using ubiquitin (UBI) as marker: T, total DNA; T+C, total DNA mixed with chloroplast DNA; T+M, total DNA mixed with mitochondrial DNA. C) Cross contamination from nuclear DNA determined by UBI presence in chloroplast and mitochondrial DNA. D) Amplification of Rubisco large subunit (RBCL) and cytochrome oxidase (COX1) in chloroplast (C) and mitochondria (M) DNA samples. P 1 : inbred line 4, P 2 : inbred line 6, H 1 : hybrid 4×6, H 2 : hybrid 6×4; L: size marker.
The steps of tissue disruption, tissue homogenization, and organelle separation by density gradient centrifugation in our miniprep are similar to other reported methods. The distinguishing feature of our method is the greatly reduced sample size and reagents quantities; we efficiently created a sucrose gradient in 2 mL tubes. Based on O’Hara-Mais and Capwell (1993), DNA extracted from chloroplast and mitochondria using our technique showed acceptable quality. So, our modifications to the cited protocols for chloroplast and mitochondria isolation rendered good quality DNA.
DNA yields obtained with these minipreps are greater than those reported by Mourad and Polacco (1988) for maize chloroplast DNA (0.1-0.2 µg DNA g-1 of sample tissue), and those recorded by Triboush et al. (1988) in organelle DNA extraction from sunflower (5-10 µg DNA g-1 tissue). While the maize protocol used 1 kg of leaves, the sunflower methodology employed between 5 and 10 g of young leaves as sample plant material. Our results show that using higher sample quantities do not necessarily produce better DNA yields. The DNA obtained throughout our minipreps has the required quality for subsequent DNA analysis.
Compared to the Kahlau et al. (2006) protocol, our chloroplast miniprep used 1 % of sample tissue, 0.05 % of extraction buffer, 0.13 % of wash buffer and 40 % of dilution buffer. Compared to the Rahman and Huber (1996) protocol our mitochondrial miniprep used 0.5 % of tissue, 0.66 % of extraction buffer and 12.5 % of wash buffer per sample (Table 3). As a result, reagent costs were reduced up to 160 times when 100 samples of chloroplast DNA extraction were processed, and up to 8 times when the same sample quantity was used for mitochondrial DNA extraction (Table 3).
Table 3 Comparison of reagents and total cost between the reference method and the method proposed in this research.
| Organelle | Protocol | Starting tissue quantity (g) | Buffer quantity (mL) | Relative cost per 100 samples (USD) | |||
| Extraction | Wash | Dilution | Lysis | ||||
| Chloroplast | Kahlau et al. (2006) | 50 | 2000 | 400 | 5 | 5 | 1 |
| Miniprep | 0.5 | 1 | 1 | 2 | 0.5 | 0.0058 | |
| Rahman and Huber (1996) | 100 | 150 | 8 | - | - | 1 | |
| Mitochondria | Miniprep | 0.5 | 1 | 1 | 2 | 0.5 | 0.125 |
PCR amplification of reference genes in DNA extracted from enriched organelle fractions was employed to identify the level of cross contamination from nuclear sources. The ubi, rbcL and cox1 PCR products coincided with the expected fragment sizes as reported by Primer3. Contamination of nuclear DNA for chloroplast DNA extraction was minimal, while mitochondrial DNA extraction showed almost no contamination (Figure 2B-C). However, as evidenced by PCR amplification of rbcL and cox1 in each enriched organelle fraction (Figure 2D), there is residual cross-contamination of each organelle type, but the quantity observed from contaminating DNA is clearly less than the DNA obtained from the enriched organelle.
Cross contamination was also reported by Bathgate et al. (1985), Diekmann et al. (2008), Gillman et al. (2007), Jo et al. (2011), Shi et al. (2012); it is attributed to organelle size plasticity from frequent fusion and division processes. It is thus difficult to obtain absolute organelle separation via density gradient centrifugation.
Conclusions
The miniprep methods proposed in this research for chloroplast and mitochondrial isolation proved reliable for DNA extraction and subsequent PCR amplification. The starting sample size is greatly minimized. These methods were tested only in samples from Manzano pepper, but little or no modification should be required to enrich organelles from other species. The methods described here have various advantages over other methods: cost reduction, proteinase disuse, percoll or cesium chloride replacement, and ultracentrifuge requirements.











 texto en
texto en 



