Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO
Share
Agrociencia
On-line version ISSN 2521-9766Print version ISSN 1405-3195
Agrociencia vol.43 n.4 Texcoco May./Jun. 2009
Recursos naturales renovables
Assessment of the origin of microbiological contamination of groundwater at a rural watershed in Chile
Determinación del origen de la contaminación microbiológica del agua subterránea en una cuenca rural en Chile
Mariela Valenzuela1* , María A. Mondaca2 , Marcelino Claret3 , Claudio Pérez3 , Bernardo Lagos4 , Oscar Parra1
1 Environmental Sciences Center EULA–Chile, University of Concepción. P. O. Box. 160–C, Concepción, Chile. *Author for correspondence: (marvalenz@udec.cl).
2 Department of Microbiology, Faculty of Biological Sciences, University of Concepción. P. O. Box. 160–C, Concepción, Chile. (mmondaca@udec.cl).
3 National Institute of Agricultural Research INIA Quilamapu, Av. Vicente Méndez 515. Chillán, Chile (mclaret@inia.cl).
4 Department of Statistics, Faculty of Physics and Mathematics, University of Concepción. P. O. Box. 160–C, Concepción, Chile.(bla@udec.cl).
Received: Febrero, 2008.
Aproved: Enero, 2009.
Abstract
In a rural watershed in Chile, the scarce groundwater available represents almost the only water source both for agriculture and domestic use. This water has microbiological quality problems, which result in an agricultural and local economic development constraint. Contamination can come from punctual or diffuse sources. Characterizing the microbiological quality of groundwater allows both to identify sources from the point of view of whether they are point or non point —thus facilitating their reduction or elimination— and to determine health hazards likely to affect the population in the area under study. This study aimed to improve the state of knowledge on the microbiological quality of groundwater at a rural watershed. Forty two wells were seasonally analyzed over a one–year period. The indicators microorganisms —total coliforms, fecal coliforms and fecal streptococci— were quantified. The study of the probable origin of the indicators was undertaken using the fecal coliform to fecal streptococci ratio, biochemical identification of enteric bacteria, and somatic coliphages detection as presence of human enteric virus indicator. Temporal contamination dynamics was determined with statistical analysis of indicator organism concentration. Results suggest that the main source of fecal contamination is of animal origin, a diffuse one. Concentrations of bacterial indicators have a temporal basis showing variable levels among seasons, with a higher concentration in the rainy one. All analyzed wells contain opportunistic pathogens.
Key words: Fecal coliforms, fecal streptococci, groundwater, microbiological contamination, total coliforms.
Resumen
En una cuenca rural en Chile, la escasa agua subterránea disponible representa casi la única fuente de agua para uso agrícola y doméstico. Esta agua tiene problemas de calidad microbiológica, que resultan en una restricción agrícola y de desarrollo económico local. La contaminación puede provenir de fuentes puntuales o difusas. La caracterización de la calidad microbiológica del agua subterránea permite tanto identificar las fuentes del punto de vista de si son puntuales o no puntuales —lo que facilitaría su reducción o eliminación— como para determinar riesgos a la salud que puedan afectar a la población del área estudiada. El objetivo de este estudio fue mejorar el estado del conocimiento acerca de la calidad microbiológica del agua subterránea en una cuenca rural. Por temporada se analizaron 42 pozos en un periodo de un año. Los microorganismos indicadores —total de coliformes, coliformes fecal y estreptococos fecales— fueron cuantificados. Para el estudio del probable origen de los indicadores, se usó la proporción de coliformes fecales a estreptococos fecales, identificación bioquímica de bacteria entérica, y la detección de colífagos somáticos como indicadores de la presencia del virus entérico humano. Se determinó la dinámica de contaminación temporal con un análisis estadístico de la concentración de organismos indicadores. Los resultados sugieren que la fuente principal de contaminación fecal es de origen animal, una fuente difusa. Las concentraciones de indicadores bacterianos tienen una base temporal, lo que conlleva a diferentes niveles entre temporadas, con una mayor concentración en la temporada lluviosa. Todos los pozos contienen patógenos oportunistas.
Palabras clave: Coliformes fecales, estreptococos fecales, agua subterránea, contaminación microbiológica, coliformes totales.
INTRODUCTION
Groundwater supply is generally perceived as less vulnerable to contamination than surface water, due to the natural filtering ability of the subsurface environment and the distance microorganisms would have to travel in order to reach the groundwater source. However, household wells in rural areas are susceptible to contamination because they are shallow, may be less carefully maintained, and can be located in close proximity to areas with loading of human or animal feces. This fecal contamination comes from different sources, livestock being the most frequent one, which creates a diffuse source (Tian et al., 2002), and inadequate on–site human waste disposal systems, which contributes to fecal contamination of watersheds as a point source. This fecal material is of high risk because of the possible presence of human pathogens (Tallon et al., 2005). The fact that contamination can come from various possible sources, makes the origin of contamination usually unknown. Therefore, knowing the origin of fecal contamination is crucial for determining associated risks; it is also vital for determining the actions needed to remediate it (Graves et al., 2002). Although quantitative methods exist, no one identifies the sources of fecal contamination in a fast and accurate manner (Bernhard and Field, 2000), nor are they always useful by themselves to make valid inferences. This occurs because this type of entries are disperse and sporadic, making their detection difficult. Indeed, fecal source identification is particularly difficult for contaminated groundwater.
Fecal coliforms and fecal streptococci are considered to originate in the digestive tract of humans and warmblooded animals. Historically, coliform bacteria have been used as water quality indicators, due to their association with the intestinal tract and with pathogenic bacteria (Conboy and Goss, 2001). Animal feces can contain pathogenic bacteria including Escherichia coli O157, Salmonella sp., Listeria monocytogenes, Campylobacter sp. (Avery et al., 2004), Shigella sp. (Leclerc et al., 2002) and Vibrio sp. However, indicator bacteria do not correlate well with the presence of viruses (Grabow, 2001). In some cases, testing of bacteriophages, mainly somatic coliphages (SOMCPH), provides additional information about fecal microorganisms reaching groundwater (Lucena et al., 2006). A coliphage is a virus that specifically infects and replicates in Escherichia coli bacteria. In terms of composition, structure and morphology, phages share many fundamental properties with human viruses. Since human enteric viruses are released into the environment almost exclusively from the gastrointestinal tract, phages which infect typical enteric bacteria such as E. coli, resemble human viruses with regard to origin and release into the environment (Grabow, 2001). In this sense, SOMCPH provide original information by tracking fecal pollution longer and further than bacterial ones (Skraber et al., 2004). Evidence of virus presence in groundwater is a concern, since viruses have a low infectious dose, and enteric viruses are the human pathogens most likely to contaminate groundwater.
In Chilean drylands, there are serious problems of water supply both for human consumption and for agricultural activities. In some watersheds, farmers obtain small amounts of water from private wells. They use groundwater sources to obtain drinking water, and for other domestic purposes, orchards, greenhouses and livestock production. Most of these farmers use water directly without any treatment and can be, therefore, exposed to a variety of water–related diseases. Improving the quality of groundwater resources is an important environmental issue for the gradual improvement of rural communitie' quality of life.
The purpose of this work was to characterize the microbiological quality of groundwater and to identify potential sources of contamination. The results will advance the understanding of dominant sources of fecal microorganisms in Chilean dryland rural watersheds, and will contribute to improve water quality.
MATERIALS AND METHODS
Study area
The small rural San José Creek Watershed (SJCW), in Chile, has an area of 10.77 km2 , and a population of approximately 60 families (Figure 1). The catchment area is diversely inhabited by families who use traditional agriculture. There is a low domestic animal density. Agricultural production consists mainly of wheat (Triticum aestivum) and lentils (Lens culinaris). The SJCW is characterized by a Mediterranean climate with a long dry season leading, consequently, to water shortage, and a short wet season. The San José Creek, which drains the watershed, has a longitude of 5.44 km with an intermittent regime. The moisture accumulation period occurs between April and June, when rains begin. Between July and October the soil is saturated, so almost all precipitation is lost as run off. From November to March precipitation is scarce and evapotranspiration is high, so soils get dry, with almost no base flow in the watershed. The average slope is 17 %, and more than half of the surface has a slope greater than 15 %. The groundwater level varies throughout the year, and wells are recharged by rain infiltration. The wells are very variable, and have an average depth of 7.0 m; a median of 5.8 m; a minimum of 1.0 m and a maximum of 19.9 m. The median domestic well yield is 1.1 L min–1 .
Water sample collection
Considering the temporal dynamics of groundwater in the area, four monitoring campaigns were carried out in a year. Groundwater quality was monitored in March, June, September and December, 2005 at 42 wells, and analyzed for microbiological quality. The sampling program was designed to gather as much information as possible in the shortest period of time. This ensured that hydrologic conditions were relatively stable, thus allowing reliable comparison between sites. Site location data were determined using global positioning system (GPS) units. The pH, specific conductance (SC), and water temperature were measured in the field. Well selection was carried out using a stratified random sample (Murray, 2002), where all wells of the watershed were classified in relatively homogeneous strata, based on information of previous monitoring on microbiological indicator level in the SJCW. Then, each stratum was randomly sampled (simple random sampling). Water samples from all sites were collected in 900 mL sterile bottles from approximately 50 cm below the water surface and transported inside coolers to the laboratory and tested within 6 h. In June, two wells could not be sampled because of terrain inaccessibility, and in December three wells could not be accessed, because permission from the well owners was denied.
Indicator organisms
The microbiological quality of water resources was determined using indicator organisms that included total coliforms, fecal coliforms and fecal streptococci. Coliform concentrations were analyzed using the membrane filtration technique, according to standard methods. Aliquots (100, 10 and 1 mL) of each water sample were filtered through a 0.45 µ Millipore membrane filter. After incubation, the number of colonies were enumerated and calculated per 100 mL–1. All samples were tested in triplicate. Results were reported as Colony Forming Units (CFU 100 mL–1). Samples that were overgrown were considered to contain > 1000 CFU 100 mL–1. Colonies forming a green metallic sheen were counted as total coliforms on m–Endo agar (Difco®, USA). For fecal coliforms counting, the filter was placed on a petri dish containing m–FC agar (Difco®, USA), which gave the selected colonies a blue color, while the selective count of fecal streptococcus was performed by incubating them in m–enterococcus agar (Difco®, USA).
Data analysis
The results for total coliforms, fecal coliforms and fecal streptococci obtained in four seasons were analyzed using the STATISTICA Statsoft Inc. software. Besides, a ratio of fecal coliforms to fecal streptococci was calculated.
Bacterial identification
Thirty water samples from three seasons (March, June and December) were inoculated in MacConkey agar. Between 1 to 6 colonies differing in size, shape, and color were randomly selected from the MacConkey agar and further isolated again on MacConkey agar and incubated at 37 °C for 24 h. Later, Gram staining and the oxidase test were performed. Isolated strains were identified according to their biochemical properties using the RapID NF and RapID ONE systems (REMEL Inc.). Preparation of organism suspensions, inoculation, incubation times and temperatures, interpretation of reactions, and quality control were performed according to the manufacturer's recommendations for each system. Strains were stored in Luria broth with glycerin at –20 °C until use.
Somatic coliphages
Somatic coliphages were enumerated by means of the plaque assay method using the double layer technique. If a phage is present in the sample, it attaches to an E. coli cell and replicates, causing the death of the cell and cell lysis, until a plaque is visible. Plaques were counted as somatic coliphages on host strain E. coli B and expressed as plaque–forming units per 100 ml (pfu/100 ml) (Grabow, 2001).
RESULTS AND DISCUSSION
Indicator organisms
Colimetries in all water samples did not meet the minimum standards for drinking water as established by the Chilean norm (0 CFU 100 mL–1). In some wells, plate counts did not comply either with the Chilean standard of 1000 CFU 100 mL–1 for irrigation purposes. Based on this criterion, a low percentage of wells cannot be used for irrigation (7.3 % of wells in March, 4.9 % in June, 0 % in September and 2.6 % in December). The most frequent indicator was total coliforms.
Considering the seasonal variations of the level of an indicator organism present in groundwater, contributes to elicit whether the sources are point or diffuse. According to the results obtained in this study, the total coliforms, fecal coliforms and fecal streptococci load in water samples seems to be seasonally regulated, with the highest concentrations occurring during winter (June), when most precipitation occurs (Figure 2). Therefore, this contamination may come from runoff, where some coliforms are associated with particles (George et al., 2004).
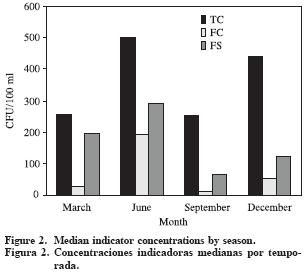
Median concentrations increase again in December. This can be due to an increase in demand from the wells during the later part of the year, combined with minimal water yield. Changes in indicator counts are assumed to reflect changes in the rate of fecal contamination of a water body. Fecal origin bacteria can survive for extended periods of time in a groundwater environment. This long survival of enteric bacteria means that groundwater has the potential to be unsafe for consumption for a long time after contamination has occurred (Conboy and Goss, 2001).
Total coliforms are ubiquitous in nature, so the results obtained may reflect that the wells are receiving allochtonous material from other possible sources like wild animals, organic material falling into the wells and soil borne bacteria, among others.
More than 50 % of the wells had a ratio <0.7, reaching 83.3 % of the samples in September. On the contrary, the ratio > 4 was found to appear in a significantly lower percentage, ranging between 0 in September and 10.3 % in December. A predominantly human source should exhibit an initially high (> 4) ratio which should then fall, whereas a non–human source should exhibit an initially low ratio (<0.7), which should subsequently rise. As used elsewhere (Donderski and Wilk, 2002; Daby et al., 2002; Troussellier et al., 2004), the ratio fecal coliforms/ fecal streptococci showed that the source of indicator bacteria is mostly animal, followed by mixed sources (Table 1).
Bacterial identification
Out of 30 well groundwater samples, a total of 123 strains were isolated. From them, 49 (39.84 %) were identified with the RapID One and RapID NF identification systems. These were considered correct if recurrent patterns of the species werehigher than 95 %. Identified bacteria are listed in Table 2. The majority of these bacteria belong to the Enterobacteriaceae family and are known as coliforms. They include E. coli, a permanent member of the intestinal community, whereas the presence of other genera like Citrobacter, Klebsiella and Enterobacter, bears a transient character (Leclerc et al., 2001). In the case of bacteria of the genus Yersinia, some have been commonly isolated from the environment, including water supplies of various types. Environmental strains, therefore, should be differentiated from some serotypes of Y. enterocolitica, which are the most frequently associated with human infections in several countries. These pathotypes can multiply in fresh waters and could constitute a major hazard to drinking water (Leclerc et al., 2002). The species identified were mainly Pseudomonads and E. coli. The Pseudomonadaceae constitute a large fraction of the bacteria found in natural waters. Although these microorganisms belong to the natural population of the water, some strains should be considered as opportunistic pathogens (Ribas et al., 2000).

The high percentage of unidentified bacteria is a result of the utilization of RapID systems, which are designed for clinical bacterial identification. From 14 isolated strains (Table 2), 13 can have an aquatic origin. Of these, Citrobacter amalonaticus, Citrobacter freundii, Citrobacter koseri, Enterobacter cloacae, Escherichia coli and Yersinia enetrocolitica may also have a fecal origin, while only one, Tatumella ptyseos, is neither fecal nor aquatic. The latter result seems very strange because Tatumella ptyseos is generally isolated from clinical samples (Farmer III et al., 1985). However, Paradis et al. (2005) analyzed the phylogeny of enterobacterial species commonly found in clinical samples, showing, for the first time, a tight phylogenetic affiliation between Pantoea and Tatumella species.
Somatic coliphages
SOMCPH were detected in two of the monitored wells for this variable. The positive detections of SOMCPH determined in the three sampling dates were 76 plaques 100 mL–1 in a well in March, and 1 plaque 100 mL–1 in a well in December. The SOMCPH limit for a low risk of viral infection is 1.00E+01 pfu 100 mL–1 (Griesel and Jagals, 2002). Therefore, the results indicate a good viral quality of well groundwater, and a low probability for the origin of fecal contamination to be human. None of these water sources posed any health risk to consumers in terms of this water quality variable. Muniesa and Jofré (2004) indicate that phage and bacteria densities and the bacterial physiological conditions needed for SOMCPH replication are rarely expected to be found in natural water environments. Therefore, it can be assumed that the detected SOMCPH come from an alochtonous source rather than from an in situ replication. However, the low level of viral contamination detected can be the result of the limited sample fraction analized. The sample with the main concentration of plaques was collected during the summer. However, whether or not SOMCPH occurrences in wells are seasonal, could not be inferred from the small numbers of viral detections.
CONCLUSIONS
The levels of indicator microorganisms in groundwater and the presence of opportunistic pathogens shows that wells are persistently contaminated and are, therefore, likely to become a health hazard for consumers. Microorganism levels appear to change substantially between seasons, each indicator having a different temporal behaviour that shows an elevated concentration of total coliforms, fecal coliforms and fecal streptococci after run off conditions in the SJCW. This indicates that rain water seems to be the main factor that determines the rate at which bacteria will move away from the source. At first glance, septic facilities were suspect of being the principal contamination sources. However, results suggest that animal sources are the most probable origin, that is, a diffuse one.
Data presented here suggest that the use of different techniques, simple as they may be, are adequate to access effective preliminary information on the microbiological groundwater quality in rural areas and the probable origin of contamination. Furthermore, access to this information can provide further protection of human health.
ACKNOWLEDGMENTS
This study was funded in partnership with Japan International Cooperation Agency JICA and carried out within the framework of the "Environmental conservation and participating rural development in mediterranean dryland of Chile" project.
LITERATURE CITED
Avery, S. M., A. Moore, and M. L. Hutchinson. 2004. Fate of Escherichia coli originating from livestock faeces deposited directly onto pasture. Letters Appl. Microbiol. 38: 355–359. [ Links ]
Bernhard, A. E., and K. G. Field. 2000. Identification of nonpoint sources of fecal pollution in coastal waters by using host–specific 16S ribosomal DNA genetic markers from fecal anaerobes. Appl. Environ. Microbiol. 66: 1587–1594. [ Links ]
Conboy, M. J., and M. J. Goss. 2001. Identification of an assemblage of indicator organisms to assess timing and source of bacterial contamination in groundwater. Water, Air, and Soil Pollut. 129: 101–118. [ Links ]
Daby, D., J. Turner, and C. Jago. 2002. Microbial and nutrient pollution of coastal bathing waters in Mauritius. Environ. Int. 27: 555–566. [ Links ]
Donderski, W., and I. Wilk. 2002. The sanitary state of water in the river Vistula between Wyszogrod and Torun. Polish J. Environ. Studies 11: 509–515. [ Links ]
Farmer III, JJ., B. Davies, W. Hickman–Brenner, A. McWhorter, G. P. Huntley–Carter, M. A. Asbury, C. Riddle, H. G. Wathen–Grady, C. Elias, and G. R. Fanning. 1985. Biochemical identification of new species and biogroups of enterobacteriaceae isolated from clinical specimens. J. Clin. Microbiol. 21: 46–76. [ Links ]
George, I., A. Anzil, and P. Servais. 2004. Quantification of fecal coliform inputs to aquatic systems through soil leaching. Water Res. 38: 611–618. [ Links ]
Grabow, W. 2001. Bacteriophages: update on application as models for viruses in water. Water SA 27: 251–265. [ Links ]
Graves, A. K., C. Hagedorn, A. Teetor, M. Mahal, A. M. Booth, and R. B. Reneau. 2002. Antibiotic resistance profiles to determine sources of fecal contamination in a rural Virginia watershed. J. Environ. Quality 31: 1300–1308. [ Links ]
Griesel, M., and P. Jagals. 2002. Faecal indicator organisms in the Renoster Spruit system of the Modder–Riet River catchment and implications for human users of the water. Water SA 28: 227–234. [ Links ]
Leclerc, H., D. A. Mossel, S. C. Edberg, and C. B. Struijk. 2001. Advances in the bacteriology of the coliform group: their suitability as markers of microbial water safety. Ann. Rev. Microbiol. 55: 201–234. [ Links ]
Leclerc, H., L. Schwartzbrod, and E. Dei–Cas. 2002. Microbial agents associated with waterborne diseases. Critical Rev. Microbiol. 28: 371–409. [ Links ]
Lucena, F., F. Ribas, A. E. Durán, S. Skraber, C. Gantzer, C. Campos, A. Morón, E. Calderón, and J. Jofré. 2006. Occurrence of bacterial indicators and bacteriophages infecting enteric bacteria in groundwater in different geographical areas. J. Appl. Microbiol. 101: 96–102. [ Links ]
Muniesa M., and Jofré J. 2004. Factors influencing the replication of somatic coliphages in the water environment. Antonie Van Leeuwenhoek Int. J. General Mol. Microbiol. 86: 65–76. [ Links ]
Murray, C. J. 2002. Sampling and data analysis for environmental microbiology. In: Hurst, C. J., R. L. Crawford, G. R. Knudsen, M. J. McInerney, and L. D. Stetzenbach (eds). Manual of Environmental Microbiology. Second Edition. ASM Press. Washington DC. pp: 166–178. [ Links ]
Paradis, S., M. Boissinot, N. Paquette, S. D. Bélanger, E. A. Martel, D. K. Boudreau, F. J. Picard, M. Ouellette, P. H. Roy, and M. G. Bergeron. 2005. Phylogeny of the enterobacteriaceae based on genes encoding elongation factor Tu and F– ATPase B –subunit. Int. J. Systematic and Evolutionary Microbiol. 55: 2013–2025. [ Links ]
Ribas, F., J. Perramon, A. Terradillos, J. Frias and F. Lucena. 2000. The Pseudomonas group as an indicator of potential regrowth in water distribution systems. J. Appl. Microbiol. 88: 704–710. [ Links ]
Skraber, S., B. Gassilloud, and C. Gantzer. 2004. Comparison of coliforms and coliphages as tools for assessment of viral contamination in river water. Appl. Environ. Microbiol. 70: 3644–3649. [ Links ]
Tallon, P., B. Magajna, C. Lofranco, and K. T. Leung. 2005. Microbial indicators of faecal contamination in water: a current perspective. Water, Air, and Soil Pollut. 166: 139–166. [ Links ]
Tian, Y. Q., P. Gong, J. D. Radke, and J. Scarborough. 2002. Spatial and temporal modeling of microbial contaminants on grazing farmlands. J. Environ. Quality 31: 860–869. [ Links ]
Troussellier, M., P. Got, M. Bouvy, M. M'Boup, R. Arfi, F. Lebihan, P. Monfort, D. Corbin, and C. Bernard. 2004. Water quality and health status of the Senegal river estuary. Marine Pollut. Bull. 48: 852–862. [ Links ]














