Introduction
Fusarium (Ascomycota, Fungi) comprises many species with a wide geographic distribution, whose divergence dates back to about 91.3 million years ago (Ma, et al., 2013), and which have a large number of biological properties (Geiser, et al., 2013). While some species are used for enzymatic preparation for industrial application, others cause serious diseases in many crops of agronomic importance (Waalwijk, et al., 2017). These fungi are also producers of mycotoxins such as trichothecenes and fumonisins, which contaminate agricultural products and make them unsuitable for consumption (Leslie, Zeller, Lamprecht, Rheeder, & Marasas, 2005). Fusarium oxysporum is probably the most commonly encountered species of Fusarium, and ranked fifth in the top 10 list of plant pathogenic fungi (Dean, et al., 2012). This soil-borne asexual fungus includes both pathogenic (plants and animals, including humans) and non-pathogenic strains that display a complex phylogenetic structure of cryptic species (Lombard, Sandoval-Denis, Lamprecht, & Crous, 2019). Isolates of F. oxysporum can cause wilting or root rot in a wide range of host plants, among which are many crops of economic importance (Gordon & Martyn, 1997). Fusarium verticillioides Sacc. Nirenberg, a fungus of ubiquitous distribution, is the most common species of Fusarium that affects corn. This hemibiotrophic species causes rotting of spikes, stems, and roots. It produces a broad spectrum of carcinogenic and teratogenic mycotoxins that reduce grain quality and affect human and animal health (Madania, Altawil, Naffaa, Volker, & Hawat, 2013; Covarelli, et al., 2012). Fusarium nygamai L.W. Burgess & Trimboli causes stem rot of the sorghum. It is also a major producer of toxins and can produce high levels of fumonisins (Leslie, Zeller, Lamprecht, Rheeder, & Marasas, 2005). Fusarium thapsinum Klittich et al. is the most important fungus causing stem rot and the mold of the sorghum grain. These diseases are common for this crop in most of the areas where it is grown; this species is morphologically very close to F. verticillioides (Summerell, et al., 2011).
Many fungi, including a large number of plant pathogens, are known to propagate clonally or only rarely undergo sexual recombination (Taylor, Hann-Soden, Branco, Sylvain, & Ellison, 2015). The sexual phase is unknown for more than 15,000 species of fungi, many of which are important phytopathogens (Arie, et al., 2000). Even in the absence of sexual recombination, fungal crop pathogens can exhibit sufficient genetic diversity to allow them to rapidly overcome new host resistances or evolve resistance against new fungicides (McDonald & Stukenbrock, 2016). Fusarium produces both sexual and asexual species where only 20% of the species have a known sexual phase (Ma, et al., 2013). In ascomycetes, the mating type locus (MAT) has a crucial role to develop the ability to mate (Kerényi, Moretti, Waalwijk, Oláh, & Hornok, 2004; Turgeon, 1998; Coppin, Debuchy, Arnaise, & Picard, 1997). By convention, the idiomorphs are called MAT1-1 and MAT1-2 (Turgeon & Yoder, 2000). The term “idiomorphic alleles” refer to the fact that these “alleles” have no significant similarity between their DNA sequences and encoded proteins, but they are in the same locus in homologous chromosomes. In heterothallic fungi, the sexual cycle is initiated when isolates of opposite mating types interact, that is, isolates that contain the idiomorph MAT1-1 with those that possess MAT1-2. In the ascomycetes, the functions of the MAT genes have generally been considered as those responsible for the regulation that governs the expression of genes involved in the mating process; this includes transcription factors involved in the expression of specific proteins that give the cell its identity with respect to the mating type (Waalwijk, et al., 2006). The composition of genes in the MAT locus can vary dramatically between species, but in filamentous ascomycetes, there are two genes that are constant: MAT1-1 always contains a gene called MAT1-1-1, which encodes a protein homologous to MAT1-1 in Saccharomyces cerevisiae. This protein has a unique motif called the α-box. MAT1-2 always contains a gene called MAT1-2-1 that encodes a protein with a DNA binding domain of the high mobility group (High Mobility Group, HMG) (Martin, Wingfield, Wingfield, & Steenkamp, 2011). In homothallic (self-fertile) ascomycetes, sexual reproduction can occur between any two individuals. These species are homologous for the MAT1-1-1 and MAT1-2-1 genes in the same genome organized in a species-specific manner, and generally more closely linked (Glass & Smith, 1994).
Molecular analysis of mating type genes is a useful tool for the research of reproductive lifestyles, as well as species relationships and research of various aspects of molecular evolution (Martin, Wingfield, Wingfield, & Steenkamp, 2011). Furthermore, knowledge of the mode of reproduction is important for the design of successful control strategies, since these are different for clonal and sexual reproductive organisms (Waalwijk, et al., 2017). The purposes of this study were: (1) to determine the type of MAT idiomorphs in Fusarium oxysporum, F. nygamai, F. thapsinum and F. verticillioides isolated from wheat and chickpea plants cultivated in several sites distributed in the central region of Mexico, (2) to estimate the frequency of mating types by species and by sites, (3) and to estimate the changes in sequences of MAT1-1 and MAT1-2 genes among and between species. It is not expected to find both idiomorphs in the same Fusarium isolate, since the four species considered here were previously reported heterothallic. Furthermore, high diversity in mating type sequences is expected due to the total number of isolates studied, the 20 sites sampled, and the provenance of the isolates from two crops.
Material and methods
Fungal isolates
Fifty-nine isolates of Fusarium oxysporum were obtained from chickpea (Cicer arietinum L.) plants with wilting and/or yellowing symptoms, cultivated in the states of Michoacán, Guanajuato and Sonora, in Mexico (Luna-Paez, Silva-Rojas, Marbán-Mendoza, & Valadez-Moctezuma, 2004). Another 51 isolates of Fusarium spp. were obtained from wheat (Triticum aestivum) plants with fusariosis and cultivated in the state of Guanajuato, Mexico (Rangel-Castillo, Valadez-Moctezuma, & Lozoya-Saldaña, 2017). These isolates (Supplementary Table S1) were maintained by regular subculture on potato dextrose agar (PDA) at 27 °C, and stored as spore suspension or mycelium in 100% glycerol at room temperature. The isolates were previously purified by hyphae tip developed in PDA medium and were incubated at 27 ºC. The fungal mycelia were collected with a spatula from the culture medium. The molecular identification of Fusarium isolates were carried out during different steps using sequences of the translation elongation factor 1-α (EF1-α; primers EF-1 5’ATGGGTAAGGA (A/G)GACAAGAC 3’ and EF-2 5’ GGA(G/A)GTACCAGT(G/C)ATCATGTT 3’ (O’Donnell, Kistler, Cigelnik, & Ploetz, 1998a)), small subunit ribosomal RNA gene (primers NMS1 5’ CAGCAGTGAGGAATATTGGTCAATG 3’ and NMS2 5’ GCGGATCATCGAATTAAATAACAT 3’ (Li, Rouse, & German, 1994)) and/or MAT genes (primers are mentioned below). Species identification (Supplementary Table S1) was made by comparison of the sequences against the databases via the Basic Local Alignment Search Tool (Blast, https://blast.ncbi.nlm.nih.gov/Blast.cgi).
DNA extraction and identification of the mating type
The total DNA of the harvested mycelium was extracted using the cetyltrimethylammonium bromide (CTAB) method, based on Doyle & Doyle (1987). The DNA quality was determined in 1% agarose gel electrophoresis and quantified by spectrophotometry (ND-1000 Thermo scientific, USA). The presence of the mating type of the 110 isolates was determined by PCR amplification using two primer pairs: FoM1-1-1 (5’ GCTTGATCTGTTCGGTCATG 3’)/FoM1-1-2 (5’ GCTGCTGCATCTTGGATTGC 3’) for MAT1-1 and FoM2-1-1 (5’ ACATATCGATAGCATCTACC 3’)/FoM2-1-2 (5’ AGGCGGTAATCTGCTGTGTA 3’) for MAT1-2 (Yun, Arie, Kaneko, Yoder, & Turgeon, 2000). The reaction mixture was composed of 200 μM of each dNTP, 1.5 U of Taq DNA polymerase (PROMEGA, Madison, WI, USA), 1× of Taq buffer, 2.5 mM of MgCl2, 100 ng of DNA, and 0.3 μM of each primer in a final volume of 25 μL. The thermocycling conditions consisted of an initial denaturation at 94 °C for 10 min, 30 cycles [94 °C for 1 min; 53 °C for 1 min; 68 °C for 2 min] and a final extension cycle of 72 °C for 10 min. The amplified fragments were separated in 1.2% agarose and visualized by UV light after staining with ethidium bromide.
Amplicon sequencing and cluster analysis
The PCR products of the MAT1-1 and MAT1-2 genes were sequenced in both directions using the same primers as in PCR reactions. To determine the genetic diversity and the changes in nucleotide bases and amino acids in the two idiomorphs (MAT1-1 and MAT1-2), 60% of the obtained PCR products were sequenced, i.e. 40 isolates with MAT1-1 and 26 with MAT1-2. By species, 51 isolates of F. oxysporum (31with MAT1-1 and 20 with MAT1-2), nine isolates of F. nygamai (three MAT1-1 and six MAT1-2), four isolates of F. thapsinum (all MAT1-1), and two isolates of F. verticillioides (both MAT1-1) were analyzed. The raw sequences were edited, and the consensus sequences were obtained using BioEdit 7.1.3.0 software (Hall, 1999). The sequences were compared and deposited (accession number is shown in Supplementary Table S1) in the GenBank database (http://www.ncbi.nlm.nih.gov/). The nucleotide sequences and their translated amino acids were aligned using the ClustalW tool implemented in MEGA6 (Tamura, Stecher, Peterson, Filipski, & Kumar, 2013). The two MAT idiomorphs were analyzed separately for cluster analysis, using nucleotide sequences or amino acid sequences as input data. The most appropriate nucleotide substitution model (Jones-Taylor-Thornton model) was determined using the tool implemented in MEGA6 (Tamura, Stecher, Peterson, Filipski, & Kumar, 2013). The Maximum likelihood and Neighbor-Joining based phylogenetic methods were employed using MEGA6, and internal branches were evaluated for 500 bootstrap replicates.
Results
Determination of mating type
The PCR products obtained with the primer pairs FoM1-1-1/FOM-1-1-2 and FoM2-1-1/FoM-2-1-2 had an approximate size of 1000 bp for MAT1-1 and 700 bp for MAT1-2. Out of the 110 isolates studied, it was found that 66 isolates contained the MAT1-1 idiomorph and 44 isolates contained the MAT1-2 idiomorph. No isolate presented the amplification of both idiomorphs (Supplementary Table S1). By crop, out of 59 Fusarium isolates from chickpea, 41 of them contained the MAT1-1 idiomorph and 18 the MAT1-2 idiomorph; while out of the 51 fungal isolates obtained from wheat, 25 contained the MAT1-1 idiomorph and 26 contained the MAT1-2 idiomorph. Regarding the distribution of Fusarium isolates across the fields where they obtained, both MAT idiomorphs were coincided in eight of the 20 sites, namely Abasolo, Celaya, Penjamo, Salvatierra, Valle de Santiago and Yuriria in the state of Guanajuato, and the localities of Morelia and Singuio in Michoacán (Table 1).
Table 1 Distribution of mating type idiomorphs in Fusarium species isolated from wheat and chickpea crops across the central region of Mexico.
| Origin | Isolates number | MAT1-1 (isoletes number and species name) | MAT1-2 (isoletes number and species name) |
|---|---|---|---|
| Abasolo, Gto. | 4 | 2 (1 Fusarium sp. and 1 F. nygamai) | 2 (1 Fusarium sp. and 1 F. nygamai) |
| Apaseo el Grande, Gto. | 1 | 1 (F. oxysporum) | 0 |
| Celaya, Gto. | 3 | 2 (2 F. oxysporum) | 1 (F. oxysporum) |
| Cortazar, Gto. | 1 | 1 (2 F. oxysporum) | 0 |
| Cuitzeo, Mich. | 4 | 4 (4 F. oxysporum) | 0 |
| El Calvario, Mich. | 3 | 3 (3 F. oxysporum) | 0 |
| INIFAP, Mich. | 1 | 1 (F. oxysporum) | 0 |
| Irapuato, Gto. | 3 | 3 (3 F. oxysporum) | 0 |
| Juventino Rosas, Gto. | 1 | 1 (F. oxysporum) | 0 |
| La purisima, Mich. | 3 | 3 (3 F. oxysporum) | 0 |
| Morelia, Mich. | 9 | 8 (8 F. oxysporum) | 1 (F. oxysporum) |
| Penjamo, Gto | 43 | 19 (8 Fusarium sp., 5 F. oxysporum, 2 F. nygamai, 2 F. thapsinum and 2 F. verticillioides) | 24 (12 Fusarium sp., 6 F. oxysporum and 6 F. nygamai, 2 F. thapsinum) |
| Puquichapio, Gto. | 2 | 2 (2 F. oxysporum) | 0 |
| Salamanca, Gto. | 4 | 4 (1 Fusarium sp., 1 F. oxysporum and 2 F. thapsinum) | 0 |
| Salvatierra, Gto. | 7 | 2 (2 F. oxysporum) | 5 (5 F. oxysporum) |
| Sinaloa, Sinaloa | 4 | 0 | 4 (4 F. oxysporum) |
| Singuio, Mich. | 5 | 4 (4 F. oxysporum) | 1 (1 F. oxysporum) |
| Valle de Santiago, Gto. | 4 | 2 (2 F. oxysporum) | 2 (2 F. oxysporum) |
| Villagran, Gto. | 1 | 0 | 1 (1 F. oxysporum) |
| Yuriria, Gto. | 7 | 4 (4 F. oxysporum) | 3 (3 F. oxysporum) |
Gto.: Guanajuato State
Mich.: Michoacán State
Variability of mating type sequences
The MAT1-1 idiomorph sequences (≈ 900 bp) consisted of two partial exons and one intron; while the MAT1-2 idiomorph (≈ 600 bp) consisted of two complete introns, two partial exons and one complete exon. The multiple sequence alignment, containing the four species of Fusarium, of the 40 nucleotide sequences of the MAT1-1 idiomorph showed the presence of 108 variable sites along the alignment (12.4% of the total alignment). Of the changes, 31.3%, 11.5% and 9.7% were found in intron, exon 1 and exon 2, respectively (Table 2). For the 31 sequences of F. oxysporum, 19 sites (2.2%) were variable from the 869 bases in the alignment. Of the changes, 2.1%, 2.3% and 1.4% were found in intron, exon 1 and exon 2, respectively (Table 2). For the two sequences of F. verticillioides, 11 sites (1.2%) were variable from the 905 bases in the alignment. Of the changes, 2.2%, 1.3% and 0% were found in intron, exon 1 and exon 2, respectively (Table 2). For the three sequences of F. nygamai, only one variable site was detected in the exon 1 along 892 bases in the alignment. Meanwhile, there was no variable site detected along the alignment of 903 bases for the four sequences of F. thapsinum.
Table 2 Genetic variability of MAT1-1 and MAT1-2 sequences from Fusarium species.
| Mating type | MAT1-1 No. variable sites / No. total sites | MAT1-2 No. variable sites / No. total sites | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Fusarium species | Total | Intron 1 | Exon 1 | Exon 2 | Total | Intron 1 | Intron 2 | Exon 1 | Exon 2 | Exon 3 |
| The four species | 108/869 | 15/48 | 86/749 | 7/72 | 44/588 | 6/56 | 5/47 | 1/23 | 21/263 | 11/199 |
| F. oxysporum | 19/869 | 1/48 | 17/749 | 1/72 | 8/631 | 3/56 | 0/47 | 0/66 | 2/263 | 3/199 |
| F. nygamai | 1/892 | 0/46 | 1/749 | 0/97 | 15/592 | 2/56 | 3/47 | 1/23 | 7/263 | 2/203 |
| F. verticillioides | 11/905 | 1/46 | 10/749 | 0/109 | - | - | - | - | - | - |
| F. thapsinum | 0/903 | 0/46 | 0/749 | 0/108 | - | - | - | - | - | - |
The multiple alignments of the 26 nucleotide sequences of the MAT1-2 idiomorph of the two Fusarium species, i.e. F. oxysporum and F. nygamai, showed the presence of 44 variable sites along the alignment (7.5% of the total alignment). Of the changes, 10.7%, 10.6%, 4.3%, 8% and 5.5% were found in intron 1, intron 2, exon 1, exon 2 and exon 3, respectively (Table 2). For the 20 sequences of F. oxysporum, eight sites (1.3%) were variable from the 631 bases in the alignment. Of the changes, 5.4%, 0%, 0%, 0.8% and 1.5% took place in intron 1, intron 2, exon 1, exon 2 and exon 3, respectively (Table 2). For the six sequences of F. nygamai, 15 sites (2.5%) were variable from the 592 bases in the alignment. Of the changes, 3.6%, 6.4%, 4.3%, 2.7% and 1% were found in intron 1, intron 2, exon 1, exon 2 and exon 3, respectively (Table 2).
Comparing the variations in the coding and non-coding regions of the MAT genes together, 17.6% of variable sites were found in the non-coding regions and 9.6% in the coding regions of the species studied. For F. oxysporum, 2.6% of variable sites were found in the non-coding regions and 1.7% in the coding regions. Meanwhile, For F. nygamai, 3.4% of variable sites were found in the non-coding regions and 0.8% in the coding regions.
The multiple sequence alignment, containing all four species of Fusarium, of the 40 amino acid sequences of the MAT1-1 idiomorph showed the presence of 42 variable residues along the alignment (15.4% of the total alignment). For the 31 sequences of F. oxysporum, 6 residues (2.2%) were variable from the 273 amino acids in the alignment. For the two sequences of F. verticillioides, 6 residues (2.1%) were variable from the 286 amino acids in the alignment. For the three sequences of F. nygamai, only one variable site was detected along 286 amino acids in the alignment. Meanwhile, there was no variable residue detected along the alignment of 281 amino acids for the four sequences of F. thapsinum. The multiple alignments of the 26 amino acid sequences of the MAT1-2 idiomorph from two Fusarium species, i.e. F. oxysporum and F. nygamai, showed the presence of 12 variable residues along the alignment (7.5% of the total alignment). For the 20 sequences of F. oxysporum, two residues (1.1%) were variable from the 175 amino acids in the alignment. For the six sequences of F. nygamai, 4 residues (2.5%) were variable from the 163 amino acids in the alignment.
Cluster analysis
The obtained trees based on the nucleotides and the translated sequences showed a similar grouping pattern. All the isolates of F. oxysporum were grouped in a single clade apart from the isolates of the species F. nygamai, F. verticillioides and F. thapsinum in the tree obtained with the sequences of MAT1-1 (Fig. 1) and from the species F. nygamai in the tree obtained with the MAT1-2 sequences (Fig. 2). The isolates F24 (from chickpea) and T8.2 (from wheat) showed some divergence, while F7 and F67 (from chickpea) showed greater divergence from the remaining isolates of F. oxysporum when analyzed with MAT1-1 (Fig. 1). The isolates F8, F57, F66 and F74 (from chickpea) were the most divergent of the remaining isolates of F. oxysporum when analyzed with MAT1-2 (Fig. 2).
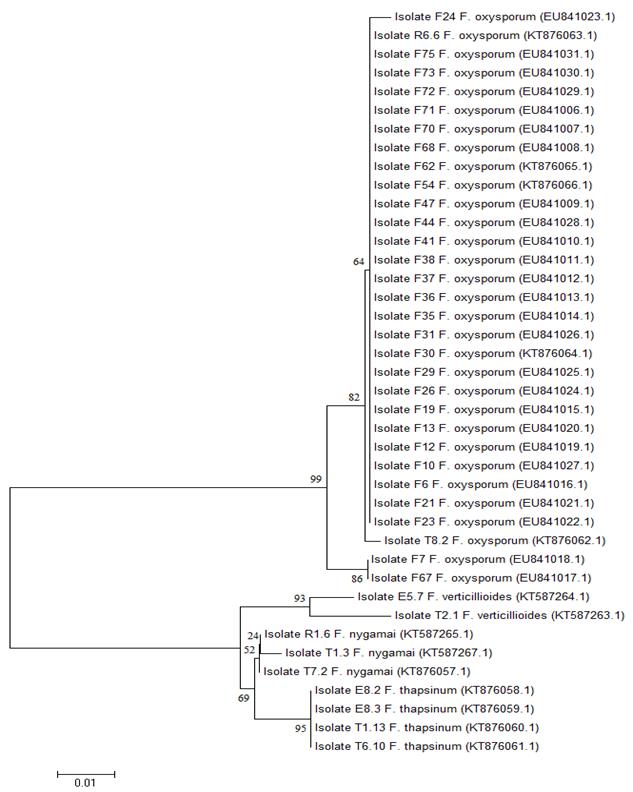
Fig. 1 Dendrogram based on amino acid sequences of the mating type MAT1-1 idiomorph for the isolates of Fusarium oxysporum, F. nygamai, F. verticillioides and F. thapsinum obtained from wheat and chickpea plants in the central region of Mexico. The Neighbor-Joining method and the Jones-Taylor-Thornton (JTT) model were applied. The numbers in the nodes represent the estimated bootstrap values from 500 repetitions. The key in parentheses indicates the reference number of the sequence in the Genbank. The isolates name initial “F” indicates isolates obtained from chickpea and the initials “E”, “R” and “T” indicates isolates obtained from wheat.
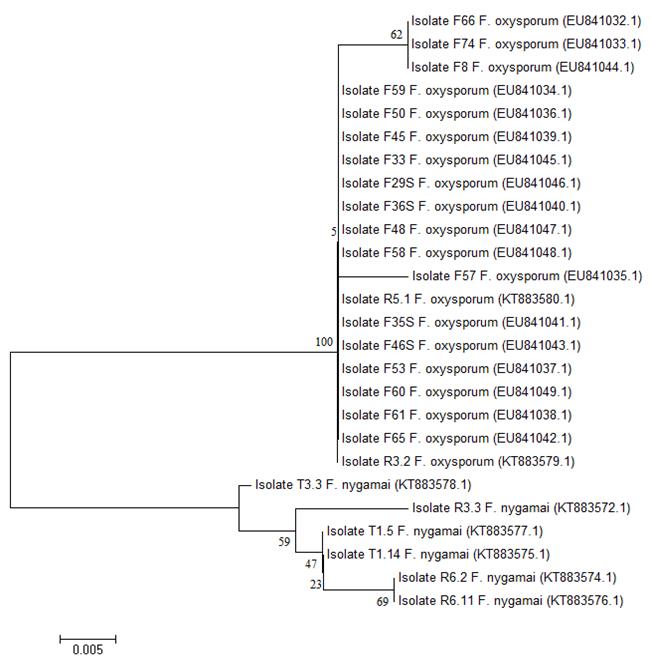
Fig. 2 Dendrogram based on amino acid sequences of the mating type MAT1-2 idiomorph for the isolates of Fusarium oxysporum and F. nygamai obtained from wheat and chickpea plants in central Mexico. The Neighbor-Joining method and the Jones-Taylor-Thornton (JTT) model were applied. The numbers in the nodes represent the estimated bootstrap values from 500 repetitions. The key in parentheses indicates the reference number of the sequence in the Genbank. The isolates name initial “F” indicates isolates obtained from chickpea and the initials “R” and “T” indicates isolates obtained from wheat.
Discussion
Using a molecular approach, it is possible to detect MAT genes also in asexual species, which is a first step towards learning the causes of asexuality (Yun, Arie, Kaneko, Yoder, & Turgeon, 2000). In the genomes of species that reproduce asexually, sequences responsible for inheritance of the type of mating were detected; even, the examined isolates of F. oxysporum and F. nygamai showed differentiation in the genetic background of the mating type. No isolate showed both idiomorphs, which is a distinctive feature of the heterothallic species. Similar results were reported for several species from the genus Fusarium (Ma, et al., 2013; Irzykowska & Kosiada, 2011; Fourie, Steenkamp, Gordon, & Viljoen, 2009; Kawabe, et al., 2005). For the species F. verticillioides and F. thapsinum, which are known to be heterothallic (Martin, Wingfield, Wingfield, & Steenkamp, 2011), only the presence of the MAT1-1 idiomorph was detected, probably due to the limited number of isolates studied, i.e. 2 and 4 isolates, respectively (species identification by sequencing).
The designation of the mating type to each Fusarium isolate is shown in Supplementary Table S1. The results revealed the presence of only one mating type in each F. oxysporum isolate, with a total of 60.8% MAT1-1 and 39.2% MAT1-2 within the isolates evaluated. Similar results were reported by Kashyap, Rai, Kumar, & Srivastava (2016) where they determined that 60% of the isolates were MAT1-1 and 40% MAT1-2 in 20 isolates of F. oxysporum f. sp. ciceris. Irzykowska & Kosiada (2011) found that from a total of 30 isolates of F. oxysporum, 33% contained MAT1-1 and 77% contained MAT1-2. While, Kawabe, et al. (2005) found that 76.7% contained MAT1-1 and 23.3% contained MAT1-2, from a total of 30 isolates of F. oxysporum f. sp. lycopersici. These values may vary depending on the number of isolates analyzed, geographic zones, and type and number of host species (Waalwijk, et al., 2006).
The multiple sequence alignments of the MAT genes of Fusarium species showed a greater divergence between species than within the same species. The four species together presented polymorphisms in 12.4% of their MAT1-1 sequences, whereas the isolates of F. oxysporum, F. verticillioides, F. nygamai and F. thapsinum varied only by 2.2%, 1.2%, 0.1%, and 0%, respectively. For MAT1-2, the isolates of F. oxysporum varied in 1.3% of their sequences and F. nygamai in 2.5%, while the divergence between the two species was higher (7.5%). These results agree with previous reports where it has been found that mating type genes are highly divergent between species (Wik, Karlsson, & Johannesson, 2008; Arie, Christiansen, Yoder, & Turgeon, 1997) and can be strongly conserved within species (Turgeon, 1998). Moreover, intra-specific variability is common in F. oxysporum despite their sexual form is unknown (Irzykowska & Kosiada, 2011; O’Donnell, Ward, Geiser, Kistler, & Aoki, 2004). Changes in non-coding regions of MAT genes in Fusarium species, namely F. oxysporum and F. nygamai were greater than in the coding regions. Martin, Wingfield, Wingfield, & Steenkamp (2011) indicated that, in general, the non-coding portions of the MAT loci are more variable among the species than the coding portions. The heterothallic MAT loci do not recombine and all the parts are strongly linked. As a result, the non-coding regions of the heterothallic MAT loci are not independent and could diverge more rapidly due to the functional restriction acting on the linked coding regions; and selection against the accumulation of deleterious mutations in MAT loci may sometimes be lacking (Clark, Aagaard, & Swanson, 2006).
Cluster analyses of the DNA and amino acid sequences derived from the coding region in the two MAT idiomorphs revealed that all the isolates of F. oxysporum were grouped together in a single clade separated from the isolates of the species F. nygamai, F. verticillioides and F. thapsinum (Figs. 1 and 2), confirming greater divergence between species than within the same species. Taxonomically, the species F. nygamai, F. verticillioides and F. thapsinum belong to the Gibberella fujikuroi species complex, African clade, a taxonomic group close to F. oxysporum complex (Geiser, et al., 2013; Martin, Wingfield, Wingfield, & Steenkamp, 2011; O’Donnell, Cigelnik, & Nirenberg, 1998b).
Although functional mating type genes have been identified (Martin, Wingfield, Wingfield, & Steenkamp, 2011; Arie, et al., 2000), the crossbreeding of F. oxysporum f. sp. lycopersici or f. sp. cubense of opposite mating types did not result in viable offspring (Fourie, Steenkamp, Gordon, & Viljoen, 2009; Kawabe, et al., 2005). Alternative mechanisms that potentially lead to genetic recombination are parasexual fusion or horizontal gene transfer. Although the mechanisms involved are not yet clear, there are examples of gene transfer across different phylogenetic boundaries at various taxonomic levels (Friesen, Faris, Solomon, & Oliver, 2008; Khaldi, Collemare, Leburn, & Wolfe, 2008). However, in the case of F. oxysporum, the exchange of genetic material through the direct transfer of genes from one fungal isolate to another would be limited by vegetative incompatibility (Lievens, Houterman, & Rep, 2009; Lievens et al., 2009). However, in an ascomycete fungus, Colletotrichum gloeosporioides, the transfer of a chromosome in laboratory conditions between vegetative incompatible isolates has been reported (He, Rusu, Poplawski, Irwin, & Manners, 1998), showing that, under certain conditions, the genetic material can be exchanged between vegetative compatibility groups.
Although many species of Fusarium are asexual (Fourie, Steenkamp, Gordon, & Viljoen, 2009; Arie, et al., 2000), the fact of determining the presence of both MAT1-1 and MAT1-2 idiomorphs from one species in the same sites namely Abasolo, Celaya, Penjamo, Salvatierra, Valle de Santiago and Yuriria in the state of Guanajuato, and the localities of Morelia and Singuio in Michoacán, increases the potential for possible genetic recombination among isolates, resulting in more aggressive and pathogenic Fusarium isolates, thus demanding more efficient strategies for crop protection.
Conclusion
Of the 110 Fusarium isolates studied, 60% of the isolates contained MAT1-1 and 40% contained the MAT1-2 idiomorph, and none isolate showed both idiomorphs, Furthermore, the nucleotides and amino acid sequences of MAT genes showed more divergence between than within species. The variation of the mating type genes and the coexistence of the two idiomorphs in the same agricultural site, point to a potential future change in the aggressiveness and pathogenicity of Fusarium isolates. These results will help to better understand the genetic diversity of some Fusarium species, especially F. oxysporum.











 nueva página del texto (beta)
nueva página del texto (beta)


