Introduction
The poinsettia (Euphorbia pulcherrima Willd. ex Klotzsch.) in its natural habitat is a shrub that blooms in winter and grows up to 3 m. The most important structural parts of this species are the bracts surrounding the flowers, due to their different sizes, shapes and colors (Canul-Ku et al., 2012a), and because of its beauty it is frequently used in landscaping (Taylor, López, Currey, & Janick, 2011). In Mexico, it can be found wild in its natural habitat, established in backyards and gardens, and cultivated as improved varieties for interior decoration (Canul-Ku et al., 2014).
This plant is native to northern Guerrero, Mexico (Trejo-Hernández et al., 2012), although it grows naturally in the tropical semi-deciduous forests of the Pacific coast, from Sinaloa, Mexico, to Guatemala, and in dry tropical forests of the Balsas Basin, particularly in northern Guerrero and Morelos (Trejo-Hernández, Olson-Zúnica, & Bye-Boettler, 2015). In 2017, 18.05 million plants were established in Mexico with a production value of 576.08 million pesos (Servicio de Información Agroalimentaria y Pesquera [SIAP], 2018), with Morelos accounting for 33.12 % of the total area established nationally, followed by Michoacán, Mexico City and Puebla (SIAP, 2018). However, the varieties available in the domestic market are produced abroad, which implies high cutting acquisition costs and agronomic management problems due to lack of adaptation (Canul-Ku et al., 2018a).
In 1950, cultivation began in Morelos of some wild poinsettia materials that differed in bract shape and color (Colinas et al., 2015), which are known as sun, backyard, heliophytic or semi-cultivated poinsettias. In recent years, there has been increased interest in these materials (Galindo-García et al., 2012) grown commercially in open-field nurseries with little technology, although information has recently been generated on their agronomic management (Galindo-García, Alia-Tejacal, Colinas-León, & Valdez-Aguilar, 2015a; Galindo-García et al., 2015b; García-Pérez et al., 2015). On the other hand, improved or shade varieties are grown in containers of different sizes and presentations for marketing during the Christmas season (Canul-Ku et al., 2014).
In the present decade, several research groups in Mexico have carried out wild and sun poinsettia collections in several states of the country (Canul-Ku, García-Pérez, Osuna-Canizales, & Ramírez-Rojas, 2012b) in order to know, conserve, protect and use this phytogenetic resource of Mexico (Galindo-García et al., 2015a), since the distribution areas of this species were reduced from one collection year to another (Canul-Ku et al., 2014). These groups have morphologically characterized 11 sun poinsettia varieties, which are registered in the Catalogo Nacional de Variedades Vegetales de México (National Catalogue of Plant Varieties of Mexico) (Colinas et al., 2015; Galindo-García et al., 2015a); in addition, Mexican varieties derived from improvement by hybridization of wild poinsettia accessions have been generated (Canul-Ku et al., 2017, 2018a; Canul-Ku, García-Pérez, Barrios-Gómez, & Rangel-Estrada, 2018b). However, these materials have not been molecularly characterized, which could serve to know their variation and genetic diversity for use in breeding programs or other activities (Canul-Ku et al., 2014).
Improved or shade poinsettias have been characterized with RAPD (Ling, Sauve, & Gawe, 1997), DAF (Starman, Duan, & Abbit, 1999) and AFLP (Parks & Moyer, 2004) markers. These techniques have been efficient in identifying poinsettia cultivars, determining relationships among cultivars and developing markers for important characteristics in closely related varieties. There are few works with molecular markers in sun and wild poinsettias (Trejo-Hernández et al. 2012, 2015; Trejo-Hernández, Briones-Dumas, Gómez-Bermejo, & Olson, 2018); however, these were done for phylogeny and evolutionary history purposes, and not to determine their diversity.
It is important to know the variability and genetic diversity of sun poinsettias in Morelos for their use and conservation, as well as to identify them, study them and use them in future research and breeding programs. Therefore, the present study was carried out to determine the genetic diversity, through RAPD markers, of 35 accessions de Euphorbia spp. collected in the state of Morelos, Mexico.
Materials and methods
The experimental trial was carried out at the Autonomous University of the State of Morelos’ Experimental Station’s Agricultural Production Laboratory, located in Cuernavaca, Morelos, Mexico. We used the RAPD molecular marker technique (Williams, Kubelik, Levak, & Tingey, 1990) and the protocol described by Doyle and Doyle protocol (1987) for DNA isolation, with some modifications recommended by Andrade-Rodríguez, Villegas-Monter, Gutiérrez-Espinosa, Carrillo-Castañeda, and García-Velázquez (2005). Thirty-five accessions collected in 19 municipalities of Morelos were studied, 34 of the species E. pulcherrima (most with red bracts, one with pink bracts and two with yellow ones) and one of E. leucocephala (with white bracts) (Table 1). Collections included four poinsettia varieties registered in the Catálogo Nacional de Variedades Vegetales del Servicio Nacional de Inspección y Certificación de Semillas (National Catalogue of Plant Varieties of the National Seed Inspection and Certification Service) (SNICS, 2018): ‘Valenciana’ (VAL), ‘Rehilete’ (REH 1), ‘Juan Pablo’ (ROSA) and ‘Amanecer Navideño’ (A. NAV) (Table 1).
Table 1 Accessions collected in different municipalities of Morelos.
| Accession | Key | Community | Municipality | Bract color | Nectary development |
|---|---|---|---|---|---|
| 1 | CUE 1 | Ocotepec | Cuernavaca | Red | Present |
| 2 | REH 1a | Santa María Ahuacatitlán | Cuernavaca | Red | Absent |
| 3 | REH 2 | Ocotepec | Cuernavaca | Red | Present |
| 4 | CUE 4 | Tetela del Monte | Cuernavaca | Red | Present |
| 5 | VALa | Tetela del Monte | Cuernavaca | Red | Absent |
| 6 | CUE 5 | Tetela del Monte | Cuernavaca | Red | Absent |
| 7 | ROSAa | Tetela del Monte | Cuernavaca | Pink | Present |
| 8 | A. NAVa | Tetela del Monte | Cuernavaca | Yellow | Present |
| 9 | PASCUA | Tetela del Monte | Cuernavaca | White | Present |
| 10 | BM 1 | Buenavista del Monte | Cuernavaca | Red | Present |
| 11 | BM 2 | Buenavista del Monte | Cuernavaca | Red | Absent |
| 12 | AMA 2 | Ocotepec | Cuernavaca | Yellow | Present |
| 13 | CUE 13 | Ocotepec | Cuernavaca | Red | Absent |
| 14 | TEP 1 | Tepoztlán | Tepoztlán | Red | Absent |
| 15 | TEP 2 | Tepoztlán | Tepoztlán | Red | Present |
| 16 | YAU | Yautepec | Yautepec | Red | Present |
| 17 | TLALT | San Miguel 30 | Tlaltizapán | Red | Present |
| 18 | PIXTL | Puente de Ixtla | Puente de Ixtla | Red | Absent |
| 19 | MIAC | Miacatlán | Miacatlán | Red | Absent |
| 20 | TET | Tetecala | Tetecala | Red | Absent |
| 21 | COATLAN | Coatlán del Rio | Coatlán del Rio | Red | Absent |
| 22 | TYC 1 | Tlayacapan | Tlayacapan | Red | Absent |
| 23 | TYC 2 | San Andrés | Tlayacapan | Red | Absent |
| 24 | TLALN | Tlalnepantla | Tlalnepantla | Red | Absent |
| 25 | TOTO | Totolapan | Totolapan | Red | Present |
| 26 | OCU | Santa Mónica | Ocuituco | Red | Absent |
| 27 | TVOL 1 | Tetela del Volcán | Tetela del Volcán | Red | Present |
| 28 | TVOL 2 | San Miguel | Tetela del Volcán | Red | Present |
| 29 | ZACU | Tlacotepec | Zacualpan | Red | Absent |
| 30 | CUA | Gabriel Tetepa | Cuautla | Red | Absent |
| 31 | JIU 1 | El Texcal | Jiutepec | Red | Present |
| 32 | JIU 2 | El Texcal | Jiutepec | Red | Present |
| 33 | TEPAL | Atotonilco | Tepalcingo | Red | Absent |
| 34 | AXO | Axochiapan | Axochiapan | Red | Absent |
| 35 | TLAQ | Las Garzas | Tlaquiltenango | Red | Absent |
a = Registered varieties.
DNA was isolated from 75 mg of tissue from fresh young leaves of five clonal plants from the 35 accessions to quantify it, amplify it and evaluate its quality. Quantification was performed in a spectrophotometer (DR5000, HACH®, Germany) using readings in two optical densities (OD). With the ratio of the absorbance reading at 260 and 280 nm (DO260/280), the purity of the nucleic acids was estimated and the DNA concentration was calculated with the following formula:
The degree of DNA integrity was determined by electrophoresis from a 2-μL aliquot of the DNA sample in ultrapure agarose gel (Invitrogen®) at 1 % (Andrade-Rodríguez et al., 2005). The electrophoresis chamber (75-2321, Apollo®, USA) operated at 80 V for 1 h; later, the obtained gel was placed in a gel documentation system (GVM20, Syngene®, United Kingdom) to be photographed on a computer (MT-M8131-35U, IBM®, USA) using the Canon® ZoomBrowser photo editor.
Random amplification of polymorphic DNA (RAPD) was performed with 21 random decapier primers (Table 2), selected from 60 previously tested indicators. The reaction mixture contained 10 μL of dNTPs, 2 μL of loading buffer (PCR buffer), 1.5 μL of magnesium chloride (MgCl), 4.7 μL of sterile deionized distilled water (sddw) and 0.3 μL (1.5 U) of native Taq® DNA polymerase enzyme (Invitrogen®). Amplification was performed using the polymerase chain reaction (PCR) technique, for which a thermal cycler (TC-412, Techne®, United Kingdom) was used with the following program: one pre-denaturation cycle at 94 °C for 4 min, 36 denaturation cycles at 94 °C for 1 min, annealing at 36 °C for 1 min and polymerization at 72 °C for 2 min, and an extension at 72 °C for 10 min (Andrade-Rodríguez et al., 2005).
Table 2 Primers used in DNA amplification.
| Primer | Base sequence |
|---|---|
| 1 | 5’ AGG GGT CTT G 3’ |
| 2 | 5’ GAA ACG GGT G 3’ |
| 3 | 5’ GGG TAA CGC C 3’ |
| 4 | 5’ CAA TCG CCG T 3’ |
| 5 | 5’ AGG TGA CCG T 3’ |
| 6 | 5’ CAA ACG TCG G 3’ |
| 7 | 5’ GTT GCG ATC C 3’ |
| 8 | 5’ GTT TCG CTC C 3’ |
| 9 | 5’ TGC TCT GCC C 3’ |
| 10 | 5’ GGT GAC GCA G 3’ |
| 11 | 5’ CTG CTG GGA C 3’ |
| 12 | 5’ GTA GAC CCG T 3’ |
| 13 | 5’ AGG GAA CGA G 3’ |
| 14 | 5’ CCA CAG CAG T 3’ |
| 15 | 5’ GTG AGG CGT C 3’ |
| 16 | 5’ CCG CAT CTA C 3’ |
| 17 | 5’ GAA CGG ACT C 3’ |
| 18 | 5’ CTC ACC GTC C 3’ |
| 19 | 5’ AAG CCT CGT C 3’ |
| 20 | 5’ CAC ACT CCA G 3’ |
| 21 | 5’ ACT TCG CCA G 3’ |
A = denine; G = guanine; T = thymine; C = cytosine.
Amplified fragments were separated by electrophoresis, with the same chamber and photograph conditions described previously, from 24 μL of the PCR reaction mixture in ultrapure agarose gel (Invitrogen®) at 1.5 % (Andrade-Rodríguez et al., 2005). The size of the fragments produced by the RAPD markers was obtained with Genetools® software.
The polymorphism percentages were obtained by dividing the number of bands generated by the polymorphic bands. The polymorphic information content (PIC) was obtained with the formula PICi = 2fi (1-fi) (Fernández, Soto, Salazar, & Betancourt, 2010; Roldán-Ruiz, Dendauw, Van Bockstaele, Depicker, & de Loose, 2000), where fi is the frequency of the bands where this presents the indicator i and the PICi is the PIC average of all the bands of said primer i. The PIC was used to determine the primers that generated more polymorphic information in the evaluated accessions.
A cluster analysis was applied using the Dice similarity coefficient (Nei & Li, 1979) and the unweighted pair group method with arithmetic mean (UPGMA), which is the most appropriate combination for this type of study according to Núñez-Colín and Escobedo-López (2011). Additionally, a resampling test was performed using the Jackknifing method (Efron, 1979), with 1,000 replicates, using the Free Tree® program ver. 0.9.1.50 (Pavlíček, Hrdá, & Flerg, 1999). Resampling methods are used to obtain numerical data on the consistency of the tree generated, and have been commonly used to decrease standard errors and confidence intervals (Mohammadi & Prassana, 2003). Subsequently, a dendrogram was drawn with the Tree View® program ver. 1.6.6 (Page, 1996) and was split into groups according to the resampling values, taking as reference the point where there was 100 % certainty.
Finally, a principal coordinates analysis was made (Gower, 2005) with the NTSYS® program ver. 2.1p. (Rohlf, 2000) to have a three-dimensional representation of the accessions and to be able to corroborate the cluster analysis groups.
Results and discussion
A total of 222 bands were revealed, of which 203 were polymorphic in the DNA amplification of the 35 accessions studied (Table 3); in addition, 100 % polymorphism was obtained in 10 primers used (1, 2, 5, 7, 11, 14, 15, 18, 20 and 21), with an average of 91.82 % (Table 3). In commercial poinsettia varieties, Ling et al. (1997) obtained 69 % polymorphism with 33 out of 48 registered polymorphic bands, well below that obtained in the present study, perhaps because they only evaluated commercial varieties; in addition, they reported that 42 out of 57 indicators were totally monomorphic. In this case, primers 8, 12 and 19 produced a greater number of fragments of Euphorbia genotypes and more than 80 % polymorphism.
Table 3 Number of fragments found, polymorphism percentage and polymorphic information content.
| Primer | Total bands | Polymorphic bands | Unique bands | Monomorphic bands | Polymorphism (%) | PIC1 |
|---|---|---|---|---|---|---|
| 1 | 8 | 8 | 2 | 0 | 100 | 0.18 |
| 2 | 12 | 12 | 5 | 0 | 100 | 0.15 |
| 3 | 12 | 9 | 4 | 3 | 75 | 0.06 |
| 4 | 10 | 9 | 2 | 1 | 90 | 0.11 |
| 5 | 9 | 9 | 3 | 0 | 100 | 0.13 |
| 6 | 10 | 8 | 2 | 2 | 80 | 0.14 |
| 7 | 11 | 11 | 1 | 0 | 100 | 0.24 |
| 8 | 15 | 12 | 4 | 3 | 80 | 0.09 |
| 9 | 9 | 7 | 2 | 2 | 77.78 | 0.06 |
| 10 | 7 | 6 | 3 | 1 | 85.71 | 0.16 |
| 11 | 9 | 9 | 0 | 0 | 100 | 0.20 |
| 12 | 14 | 12 | 3 | 2 | 85.71 | 0.18 |
| 13 | 11 | 9 | 2 | 2 | 81.82 | 0.20 |
| 14 | 10 | 10 | 4 | 0 | 100 | 0.18 |
| 15 | 11 | 11 | 2 | 0 | 100 | 0.19 |
| 16 | 9 | 8 | 2 | 1 | 88.89 | 0.20 |
| 17 | 11 | 10 | 1 | 1 | 90.91 | 0.11 |
| 18 | 12 | 12 | 4 | 0 | 100 | 0.12 |
| 19 | 13 | 12 | 2 | 1 | 92.31 | 0.21 |
| 20 | 8 | 8 | 0 | 0 | 100 | 0.30 |
| 21 | 11 | 11 | 4 | 0 | 100 | 0.10 |
| Total | 222 | 203 | 52 | 19 | - | - |
| Mean | 10.57 | 9.67 | 2.48 | 0.90 | 91.82 | 0.16 |
1PIC = polymorphic information content.
The total number of amplified bands ranged from 7 to 15, with an average of 10.5 bands per primer (Table 3). This value was higher than the 5.4 bands, on average, reported in shade poinsettias (Ling et al., 1997). The average PIC obtained was 0.16, with primer 20 obtaining the highest value, followed by 6 and 19 (Table 3), which indicates the discriminatory power of these markers when differentiating the genotypes studied, so its use is recommended in future studies of Euphorbia. Likewise, 52 unique bands were identified, of which PASCUA presented 46, ROSA and JIUT 1 two, and TVOL 1 and TVOL 2 only one (Table 2).
High genetic variability was detected among and within the species studied according to their polymorphism percentages and PIC values. In addition, the PASCUA, TVOL 2, ROSE, AMA 2, JIU 1 and JIU 2 accessions showed greater polymorphism in their obtained DNA fragments, as well as a greater number of unique fragments (Figure 1).
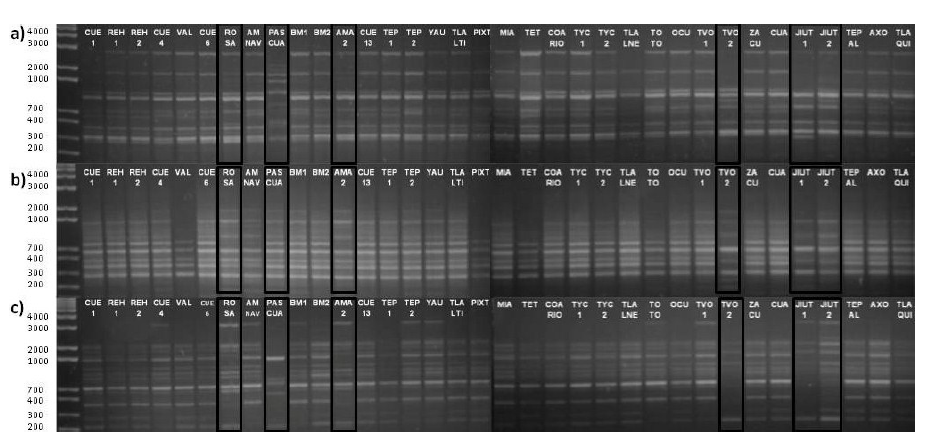
Figure 1 DNA amplification of accessions collected in Morelos, Mexico, in 1.5 % agarose gel stained with ethidium bromide. Only three primers of the 21 used are shown: a) 8, b) 13 and c) 19.
From the cluster analysis, seven groups were formed with 100 % consistency and genetic differences among them, which indicates a wide genetic diversity of the accessions collected in Morelos (Figure 2). The group with the highest number of integrated accessions was 2 with 21, followed by group 1 with eight, group 5 with two and the rest of the groups with only one accession, which indicates the differences presented (Figure 2).

Figure 2 Molecular characterization dendrogram, by means of the Dice similarity coefficient, of 35 accessions collected in Morelos, Mexico.
According to Mohammadi and Prasanna (2003), percentages greater than 70 in the internal branches of the dendrogram are acceptable. In this case, values higher than 93 % were obtained, and in 18 branches 100 % was obtained, values adequate for the formation and consistency of the created groups (Tapia-Pérez & Legaria-Solano, 2007) (Figure 2). The proportions of the internal branches of the dendrogram can be used to demonstrate the consistency of the groups, as has been used in some crops: Jatropha curcas L. (Pecina-Quintero et al., 2011), sesame (Sesamuvm indicum L.) (Salazar, Laurentin, Dávila, & Castillo, 2006) and guava (Psidium guajava L.) (Tapia-Pérez & Legaria-Solano, 2007), to characterize molecularly or identify genetic variability. In addition, these values are useful to determine the cut of the dendrogram, taking into account the branches that present 100 %.
Group 1 was characterized by the presence of sun poinsettia plants that developed nectaries, except the PIXT accession (Figure 2 and 3a). All accessions of this group had red bracts, except A. NAV (pink bracts) which is a variety registered in Mexico, suggesting that the genetic content is similar in the others (Figure 2).
The plants of group 2 presented red bracts, in addition to grouping two registered accessions (REH 1 and VAL). The REH 1 and REH 2 accessions have deformed bracts, so they probably arose as a modification of the ‘Valenciana’ poinsettia (Figure 3b). The 'Valenciana' and 'Rehilete' sun varieties are the most cultivated in Morelos (Galindo-García et al., 2012), which is why this group is the most numerous.
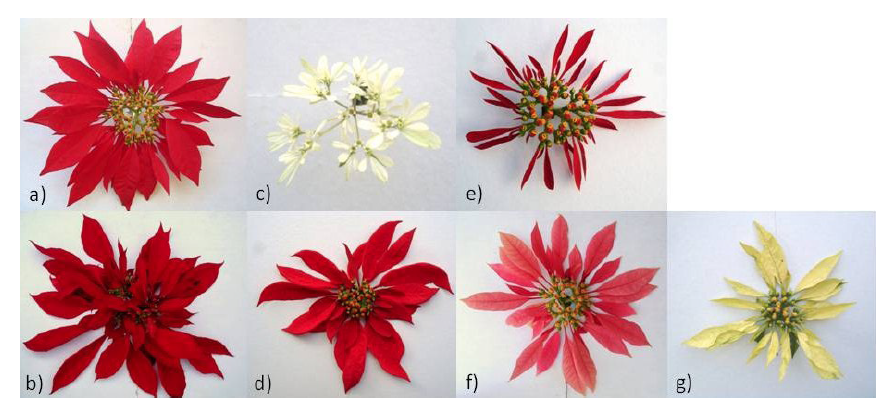
Figure 3 Examples of accessions that showed the greatest genetic diversity of each group: a) group 1 (TEP 2), b) group 2 (VAL), c) group 3 (PASCUA), d) group 4 (TVOL 2), e) group 5 (JIU 1), f) group 6 (ROSA) and g) group 7 (AMA 2).
Groups 1 and 2 presented collected materials and registered varieties of sun poinsettia, so it can be inferred that the latter are the result of domestication of species due to the transport of plants from one place to another, since the first settlers in the large cities dispersed to other municipalities and took this plant material (Trejo-Hernández et al., 2015), although another option is that the inhabitants obtained the plant material from wild populations to decorate their gardens (Trejo-Hernández et al., 2018).
Six pairs of accessions in group 2 were very similar genetically to each other: TYC 1 and 2, REH 1 and 2, TET and VAL, BM 2 and CUE 13, AXO and TEP, MIAC and CUE 6. The CUE 1 accession is close to REH 1 and 2, and TLAQ is close to AXO and TEP 1, possibly due to the gene flow reported by Tapia-Pérez and Legaria-Solano (2007). These authors state that the similarity among accessions can be caused by two factors: transport of seeds by birds and transport of seeds or cuttings of genotypes by producers to propagate them. This paper suggests that inhabitants of other municipalities bought sun poinsettia plants in Cuernavaca and took them back to their places of origin. Trejo-Hernández et al. (2015) state that there are no known natural hybrids of this plant, so it is suggested that the dispersion of the sun poinsettia has been by cuttings.
Group 3 consisted of only the PASCUA accession (E. leucocephala) collected in Cuernavaca, which showed nectary development and very small white bracts (Figure 3c). This accession belongs to another species, which coincides with it having the greatest number of different DNA fragments, separating it from the others in the dendrogram (Figure 2). Likewise, group 4 was also formed by one accession (TVOL 2) (Figure 3d), which has red bracts, nectary development and a slight deformation of bracts, so it is inferred that it may be a wild material, since it was collected in a natural habitat in Tétela del Volcán. This corroborates the result obtained with TVOL 2, which was genetically different from the others, and presented a unique fragment and a fragment that only coincided with one of PASCUA.
Group 5 consisted of two accessions: JIU 1 and JIU 2 (Figure 3e), with red bracts and nectary development, both from Texcal, Jiutepec, Morelos, and wild habitat. In Jiutepec there is an important population of Euphorbia pulcherrima in an ecological conservation area decreed in 1992, currently declared as a protected natural area (Comisión Estatal del Agua y Medio Ambiente [CEAMA], 2018). Trejo-Hernández et al. (2015) indicate that apparently wild plants develop simple inflorescences with genetic variants. In JIU 1 and JIU 2, three unique fragments were identified, i.e., they did not appear in other accessions, which placed them in a different group.
Group 6 was formed by the ROSE genotype (Figure 3f), which has pink bracts and nectary development, giving rise to simple inflorescences. Finally, group 7 consisted of the AMA 2 accession (Figure 3e), which develops nectaries and thin yellow bracts with slight deformation. It is suggested that its habitat is natural because it is located within the city limits of Cuernavaca and shows similarity with ROSA, since they coincide in having two fragments only presented by both genotypes.
The genetic variability of the accessions originating in Morelos is mainly attributed to the collection site, since the wild plants showed differences in their genotype, which implies that varietal purity is preserved (Fernández et al., 2010). Another correlation variable was bract color, where most of the accessions from different municipalities in Morelos are red and those of different color were concentrated in Cuernavaca: PASCUA (white), AMA 2 and A. NAV (yellow), and ROSE (pink); in addition, the registered varieties were grouped according to bract color: ‘Amanecer Navideño’ (A. NAV-yellow) in group 1, ‘Valenciana’ and ‘Rehilete’ (VAL and REH-red) in group 2, and ‘Juan Pablo’ (ROSA-pink) in group 6. This coincides with Prasad (2014), who reported that the color of the flowers in Hibicus rosasinensis determines the grouping of accessions.
The grouping of the 35 accessions collected in Morelos was shown to be consistent in the seven groups well-defined in the cluster analysis and represented in the principal coordinates analysis plot (Figure 4), which were differentiated by habitat and bract color. In this three-dimensional scatter plot, one can notice the distance of PASCUA (group 3) with the others, as it is a different species.

Figure 4 Three-dimensional scatter plot, obtained by principal coordinates analysis, of the 35 accessions collected in Morelos, Mexico.
Nectary development was found in 16 accessions (CUE 1, REH 2, CUE 4, ROSA, A. NAV, AMA 2, PASCUA, BVM 1, TEP 2, YAU, TLALTI, TOTO, TVOL 1, TVOL 2, JIU 1 and JIU 2), which is of utmost importance due to the possible crossing between plants. The rest of the accessions did not have nectaries, so they did not develop seed and instead sprouted some structures known as bracteoles (Colinas et al., 2015). These accessions cannot be used in crosses because they are present in all the groups described here. Identifying and crossing genetically distant genotypes increases the probability of having heterosis in the system (Wright, 1978).
The analysis of genetic diversity allowed obtaining a reliable grouping of the accessions collected in Morelos. In addition, groups of core accessions were identified with the possibility of using them in future breeding studies (Mohammadi & Prasanna, 2003; Valera-Montero, Hernández-Dávila, Silos-Espino, & Flores-Benítez, 2017).
Conclusions
Molecular characterization allowed the detection of a wide genetic diversity of accessions collected in Morelos, Mexico, with important characteristics to be used in future breeding programs. With RAPD molecular markers and statistical analyses, the collected accessions were differentiated and grouped, identifying 203 polymorphic fragments (91.82 %) that determined the degree of genetic diversity in the study population. Seven consistent groups were formed, which were associated with their bract color and habitat. The accessions that presented the greatest variability in their DNA fragments were PASCUA, TVOL 2, JIU 1, JIU 2, ROSA and AMA 2. The species E. leucocephala was different from E. pulcherrima in its genotype.











 texto en
texto en 


