Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Revista de la Sociedad Química de México
versión impresa ISSN 0583-7693
Rev. Soc. Quím. Méx vol.46 no.3 Ciudad de México jul./sep. 2002
Investigación
Two Notably Similar Proteins Follows Different Unfolding Pathways
María Elena Chánez-Cárdenas1 and Edgar Vázquez-Contreras2*
1 Departamento de Bioquímica. Instituto de Fisiología Celular, Universidad Nacional Autónoma de México. 04510, México, D.F.
2 Instituto de Química. Departamento de Bioquímica. Universidad Nacional Autónoma de México. Circuito Exterior, Ciudad Universitaria México, D.F. 04510, México. Tel: (+52) (55) 5622-4565; Fax: (+52) (55) 5616-2217. E-mail: vazquezc@servidor.unam.mx
Recibido el 15 de enero del 2002.
Aceptado el 12 de junio del 2002.
Abstract
The amino acid sequences and the three dimensional structures of triosephosphate isomerase from the parasites Trypanosoma brucei and Trypanosoma cruzi, are markedly similar. In spite of these structural similarities, we found that the equilibrium unfolding pathway of both enzymes induced by guanidinium hydrochloride is different, as showed by alterations in intrinsic fluorescence of tryptophan residues. This behavior is possibly related with the different reactivity that these enzymes presents against derivatizing cysteine residues agents.
Keywords: Triosephosphate isomerase, three dimensional structures, proteines, unfolding pathway, fluorescence.
Resumen
La composición de aminoácidos y la estructura tridimensional de la triosafosfato isomerasa de los parásitos Trypanosoma brucei y Trypanosoma cruzi son muy similares. A pesar de estas semejanzas estructurales, encontramos que la vía del desplegamiento al equilibrio inducido por clorhidrato de guanidinio y seguida por alteraciones en la fluorescencia intrínseca de los residuos de triptofano, es diferente para ambas enzimas. Este comportamiento está posiblemente relacionado con la diferente reactividad que estas enzimas presentan contra agentes derivatizantes de residuos de cisteína.
Palabras clave: Triosafosfato isomerasa, estructura tridimensional, proteinas, desplegamiento, fluorescencia.
Dedicated to Dr. Barbarín Arreguín Lozano
Introduction
The fifth reaction of glycolysis is catalyzed by triosephosphate isomerase (TIM). The catalytic properties of this enzyme have been extensively studied and it is considered a perfect catalyst [1]. TIM is a homodimeric enzyme, each monomer is a typical ( β / α)8 barrel formed by approximately 250 residues and the catalytic site is near the interface between subunits. The oligomeric state of this enzyme depends on non-covalent interactions between the monomers and is required for catalysis [2-4].
Structural information for different TIM sources is available, among them are the Trypanosoma brucei (TbTIM) [5], Trypanosoma cruzi (TcTM) [6], Saccharomyces cerevisiae (yTIM) [7], multicellular eukaryotes including human (hTIM) [8], and several prokaryotes including both bacterial and archaeobacterial groups [9-12]. This information shows that all TIMs are homodimers, with the exception of Methanothermus fervidus and Pyrococcus woesei TIMs, which are homotetramers [13].
Although the tertiary structure of TIM has been conserved, there are many differences in the primary structure of the protein from different species, and surprisingly, the catalytic efficiency of the enzyme has not been affected by these changes. The analysis of the conservation or substitution of residues in the TIM enzyme are of interest to understand the different behavior of homologous proteins in stability, susceptibility to inhibitor reagents, catalytic efficiency and folding pathways, among others.
In this work, we report the unfolding pattern of two practically identical TIMs, those from the trypanosomatids parasites T. cruzi and T. brucei. These glycosomal enzymes [14, 15] have a high identity in their aminoacid sequence and catalytic properties [6, 16]. Besides the catalytic and structural similarities, these two enzymes show great differences in the susceptibility of the interface C residue (Cys 14 and 15 respectively) to sulfhydryl reagents [6]. Since TbTIM and TcTIM have their W residues in equivalent positions, it is interesting to follow the intrinsic fluorescence of the enzymes along their denaturation pattern in order to obtain information of the conservation or not of the unfolding behavior in two homologous homodimeric enzymes.
Results and Discussion
Comparison of the primary structure of TbTIM and TcTIM
The similarity between the primary structure for TbTIM and TcTIM is shown in the Fig. 1.
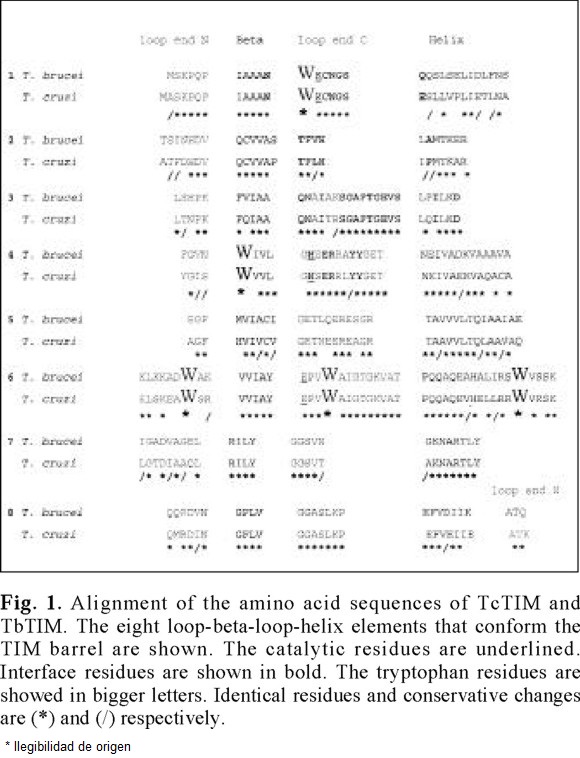
Of the comparison of the two sequences, the following data are obtained easily: as for all others TIMs analyzed to date, the catalytic residues are strictly conserved. Of the 250 residues that are possible to compare, (TbTIM possess 251), 185 i.e. 74 %, are identical [6, 16]. The remaining 26 % of the sequences present 30 conservative changes, which indicates that the enzymes possess only 13 % of non-conservative changes.
The hydrophobic, aromatic and fluorescent tryptophan residues occupy the same positions in both enzymes (12, 90, 159,170 and 193, TbTIM numeration). Considering that these enzymes present different behavior to the catalytic inactivation with derivatizing agents of cysteine residues [17], and taking into account that they are homologous proteins, their denaturation pattern induced by guanidinium hydrochloride (Gdn-HCl) was investigated. Despite this differences both enzymes shows identical native structural folds [6].
Equilibrium time
The future intention of the study of these enzymes is to analyze the energetic changes involved in the formation of the homodimeric TIM. The changes along time in the wavelength of maximal emission (λmax), spectral center of mass (SCM) and fluorescence intensity at native max (FI) of samples equilibrated in the pre-, post- and denaturing concentrations of Gdn-HCl, indicated that for both enzymes, an adequate time for equilibrium measurements is 48 h (data not shown). We found that after this period all measured signals had a minimum variation with respect to time.
Changes in tertiary structure
After equilibrium incubation of the homodimers, Gdn-HCl induces a red shift in SCM and λmax for both enzymes. There was a slightly difference between TbTIM and TcTIM: the native λmax after excitation at 295 nm of the former enzyme was 322 nm and for the latter is 317. 4 nm. Considering the same position for the W residues in both enzymes (Fig. 1), this difference in λmax possibly reflects that at least one of these intrinsic fluorophores is more buried in TcTIM that in TbTIM; for example in the yTIM, which possess only three tryptophan residues (89, 158 and 169), the λmax of the native enzyme is 322 nm after excitation at 295 nm [18]. In contrast, at high denaturant concentrations the λmax for both Tc and TbTIM are 350 nm, indicating complete exposure of tryptophan residues and the presence of completely denaturated monomers. The increase of denaturant concentration induces a decrease in fluorescence intensity at native λmax in both enzymes.
To compare the unfolding profiles of both enzymes, the apparent fraction of native dimers (xN) as a function of Gdn-HCl concentration (x) was calculated with equation (1):

where Y(x) is the change in the spectroscopic property (λmax, SCM or FI) at different x values, and YNand YUare the specific spectroscopic values for native homodimer and unfolded monomer respectively. The result of this analysis is shown in Fig. 2.
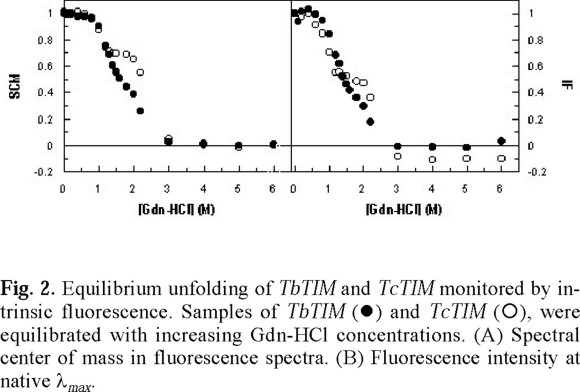
The analysis of the unfolding pattern of TbTIM and TcTIM shows that the overall transition from the native homodimer to the unfolded monomers occurs in the same interval of Gdn-HCl concentrations i.e. at concentrations higher than 3.0 M of denaturant the SCM and the FI remains constant (Fig. 2), indicating that the exposition of the tryptophan residues is not longer affected by the increase in denaturant concentration. In these conditions, there is a complete exposure of tryptophan residues and completely denaturated monomers are obtained. In the range between 0-1.0 M Gdn-HCl, both SCM and FI remain constant with values similar to those observed in the native enzyme (Fig. 2). The main difference in the unfolding pattern between these enzymes is shown in the interval 1.0-2.0 M of denaturant (Fig. 2). In this range, TbTIM follows the typical monophasic behavior found in the human [4], yeast [18, 19], Plasmodium falciparum [20] and Leshmania mexicana [21] enzymes.
On the other hand, in the same range of Gdn-HCl concentration, TcTIM stabilizes as an intermediate (Fig. 2). This same range of Gdn-HCl concentration is also populated by a monomeric intermediate in the unfolding of yTIM [18, 19]. However, in this enzyme, the transition is monophasic when the change in secondary as well as tertiary structure is verified [18]. In the unfolding of P. falciparum TIM, another intermediate was found in a near region, but characterized as an aggregate [20].
TIM unfolding and refolding from several species has been studied extensively [2, 4, 18-27]. Thermodynamic data for unfolding have been reported only for human and rabbit enzymes [4, 26, 27], where a three-state model (N ↔ 2M ↔ 2U) has been proposed. In hTIM, the unfolding of the wild-type enzyme was modeled as a two-state transition and information on the properties of the monomer was obtained from the two-state unfolding of monomeric mutants [4, 27], or calculated from equilibrium and activation energies for the wild-type enzyme [26]. The same unfolding behavior was found with yTIM [18, 19], the first experimental evidence of an equilibrium intermediate in the unfolding of the triosephosphate isomerase. In the present work we found that the transition between the homodimeric enzyme to the unfolded monomers of TcTIM also possess an equilibrium intermediate which is not clearly detectable in their homologous protein TbTIM. This difference is rather surprising taking into account the high similarity found in the aminoacid sequence, although clearly reminds the differences showed by these enzymes to the inhibition by derivatizing cysteine residues [17].
Equilibrium and kinetic folding pathways of several homologous proteins have been studied. At the beginning early studies concluded that the folding routes of homologous proteins follow fundamentally similar paths and that the folding of a certain conformation is conserved throughout evolution [28-31]. However, there are few examples of homologous proteins which unfolds in different patterns e.g. Im7 and Im 9 [32], and human Stefin A and Stefin B where the former follows a two-state pattern and the latter shows a "molten globule-like" intermediate [33]. Regarding TIM, equilibrium unfolding studies with enzymes from different sources, have shown a two-state behavior with urea [4, 18, 27, 20] or Gdn-HCl [21] as denaturant. In other studies, equilibrium-folding intermediates have been detected in the Gdn-HCl induced unfolding [18, 19, 20] or at extreme pH values [21]. All of these equilibrium intermediates were apparent when tryptophan fluorescence was followed as a function of denaturant concentrations. Here we report the presence of an intermediate state in the denaturation of TcTIM, which possess different fluorescence properties from those observed in the native or unfolded enzyme and that is not present in the unfolding of a highly similar homologous protein TbTIM.
Additional experiments to explore the origin for the differences in the unfolding behavior between these enzymes and to determine the changes in free energy of homodimer dissociation and monomer unfolding TIM are currently underway.
Experimental
Overexpression and purification of recombinant proteins
BL21(DE)3 pLysS strain expressing TbTIM was grown at 37 °C in Luria-Bertani medium supplemented with 100 µg mL-1 ampicillin and 34 µg mL-1 chloramphenicol. Culture was induced at A600 = 0.8-1.0 by addition of isopropyl-β-D-thiogalactopyranoside (IPTG) at a final concentration of 0.4 mM. Growth was continued overnight. The purification of TbTIM was performed as described in [22]. The overexpression and purification of TcTIM were performed as described [16].
Gdn-HCl unfolding experiments monitored by fluorescence
Denaturation experiments were performed incubating 50 µg mL-1 of Tc or TbTIM at 25 °C in 100 mM TEA /10 mM EDTA /1.0 mM DTT, pH 7.4 for at least 48 hours. Gdn-HCl concentration was increased from 0 to 6.0 M. The fluorescence changes were performed using an ISS PCI Photon Counting Spectrofluorometer (ISS, Urbana, IL) at 25 °C using an excitation wavelength of 295 nm (slit width = 1 mm). The spectral center of mass (SCM) of each spectrum defined as
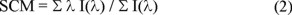
was calculated with the ISS software. In equation (2) I(λ) is the fluorescence intensity at wavelength λ. Reference samples at each denaturant concentration were subtracted from all the experimental measurements.
Acknowledgments
We thank Laboratorio de Fisicoquímica y Diseño de Proteínas, Facultad de Medicina, UNAM for loan the equipment used in this work. We are grateful to Alfredo Téllez for his help in TcTIM purification. We also like to thank Dr. Barbarín Arreguín and Dr. Enrique García-Hernández for critically reading the manuscript.
References
1. Knowles, J.R.; Albery, W.J. Accounts Chem. Res. 1977, 10, 105-111. [ Links ]
2. Zabori, S.; Rudolph, R.; Jaenicke, R. Z. Naturforsch. 1980, 35c, 999-1004. [ Links ]
3. Borchert, T.V.; Zeelen, J. Ph.; Schliebs, W.; Callens, M.; Minke, W.; Jaenicke, R.; Wierenga, R.K. FEBS Lett. 1995, 367, 315-318. [ Links ]
4. Mainfroid, V.; Terpstra, P.; Beauregard, M.; Frère J.M.; Mande, S.C.; Hol, W.G.J.; Martial, J.A.; Goraj, K. J. Mol. Biol. 1996, 257, 441-456. [ Links ]
5. Wierenga, R.K.; Noble, M.E.M.; Vriend, G.; Nauche, S.; Hol, W.G.J. J. Mol. Biol. 1991, 220, 995-1015. [ Links ]
6. Maldonado, E.; Soriano-García, M.; Moreno, A.; Cabrera, N.; Garza-Ramos, G.; Tuena de Gómez-Puyou, M.; Gómez-Puyou, A.; Pérez-Montfort, R. J. Mol. Biol. 1998, 283, 193-203. [ Links ]
7. Lolis, E.; Alber, T.; Davenport, R.C.; Rose, D.; Hartman, F.C.; Petsko, G. Biochemistry 1990, 29, 6609-6618. [ Links ]
8. Mande, S.C.; Mainfroid, V.; Kalk, K.H.; Goraj, K.; Martial, J.A.; Hol W.G.J. Prot. Sci. 1994, 3, 810-821. [ Links ]
9. Delboni, L.F.; Mande, S.C.; Rentier-Delrue, F.; Mainfroid, V.; Turley, S.; Vellieux, F.M.D.; Martial, J.A.; Hol, W.G.J. Protein Sci. 1995, 4, 2594-2604. [ Links ]
10. Alvarez, M.; Zeelen, J.P.; Mainfroid, V.; Rentier-Delrue, F.; Martial, J.A.; Wyns, L.; Wierenga, R.K.; Maes, D. J. Biol. Chem. 1998, 273, 2199-2206. [ Links ]
11. Maes, D.; Zeelen, J.P.; Thanki, N.; Beaucamp, N.; Alvarez, M.; Dao Thi, M.H.; Backmann, J.; Martial, J.A.; Wyns, L.; Jaenicke, R.; Wierenga, R.K. Proteins 1999, 37, 441-453. [ Links ]
12. Walden, H.; Bell, G.S.; Russell, R.J.M.; Siebers, B.; Hensel, R.; Taylor, G.L. J. Mol. Biol. 2001, 306, 745-757. [ Links ]
13. Kohlhoff, M.; Dahm, A.; Hensel, R. FEBS Lett. 1996, 383, 245-250. [ Links ]
14. Opperdoes, F.R.; Borst, P. FEBS Lett. 1977, 80, 360-364. [ Links ]
15. Opperdoes, F.R.; Baudhuin, P.; Coppens, I.; De Roe, C.; Edwards, S.W.; Weijers, P.J.; Misset, O. J. Cell Biol 1984, 98,1178-1184. [ Links ]
16. Ostoa-Saloma, P.; Garza-Ramos, G.; Ramírez, J.; Becker, I.; Berzunza, M.; Landa, A.; Gómez-Puyou, A.; Gómez-Puyou, M. T.; Pérez-Montfort, R. Eur. J. Biochem. 1997, 244, 700-705. [ Links ]
17. Garza-Ramos, G.; Cabrera, N.; Saavedra-Lira, E.; Gómez-Puyou, M.; Ostoa-Saloma, P.; Pérez-Montfort, R.; Gómez-Puyou, A. Eur. J. Biochem. 1998, 253, 684-691. [ Links ]
18. Vázquez-Contreras, E.; Zubillaga, R.A.; Mendoza-Hernandez, G.; Costas, M.; Fernandez-Velasco, D.A. Prot. Pept. Lett. 2000, 7, 57-64. [ Links ]
19. Morgan, C.J.; Wilkins, D.K.; Smith, L.J.; Kawata, Y.; Dobson, C.M. J. Mol. Biol. 2000, 300, 11-16. [ Links ]
20. Gokhale, R.S.; Ray, S.S.; Balaram, H.; Balaram, P. Biochemistry 1999, 38, 423-431. [ Links ]
21. Lambeir, A.M.; Backmann, J.; Ruiz-Sanz, J.; Filimonov, V.; Nielsen, J.E.; Kursula, I.; Norledge, B.V.; Wierenga, R.K. Eur. J. Biochem. 2000, 267, 2516-2524. [ Links ]
22. Borchert, T.V.; Abagyan, R.; Jaenicke, R.; Wierenga, R.K. Proc. Natl. Acad. Sci. USA, 1994, 91, 1515-1518. [ Links ]
23. Schliebs, W.; Thanki, N.; Jaenicke, R.; Wierenga, R.K. Biochemistry, 1997, 36, 9655-9662. [ Links ]
24. Fernández-Velasco, D.A.; Sepúlveda-Becerra, M.; Galina, A.; Darszon, A.; Tuena de Gómez-Puyou, M.; Gómez-Puyou, A. Biochemistry, 1995, 34, 361-369. [ Links ]
25. Waley, S. Biochem. J. 1973, 135, 165-172. [ Links ]
26. Rietveld, A.W.M.; Ferreira, S. Biochemistry, 1998, 37, 933-937. [ Links ]
27. Mainfroid, V.; Mande, S.C.; Hol, W.G.J.; Martial, J.A.; Goraj, K. Biochemistry, 1996, 35, 4110-4117. [ Links ]
28. Hollecker, M.; Creighton, T.E. J Mol Biol 1983, 168, 409-437. [ Links ]
29. Krebs, H.; Schmid, F.X.; Jaenicke R. J Mol Biol 1983, 169, 619-635. [ Links ]
30. Stackhouse, T.M.; Onuffer, J.J.; Matthews, R.; Ahmed, S.A.; Miles, E.W. Biochemistry 1988, 27, 824-832. [ Links ]
31. Kragelund, B.B.; Hojrup, P.; Jensen, M.S.; Schjerling, C.K.; Juul E.; Knudsen, J.; Poulsen, F.M. J Mol Biol 1996, 256, 187-200. [ Links ]
32. Ferguson, N.; Capaldi, A.P.; James, R.; Kleanthous, C.; Radford S.E. J Mol Biol 1999, 286, 1597-1608. [ Links ]
33. Zerovnik, E.; Virden, R.; Jerala, R.; Turk, V.; Waltho, J.P. Proteins 1998, 32, 296-303. [ Links ]




![A Non-sterically Preferred Conformation in Bis-1,4-(2-methyl-4, 5-dihydro-1H-benzo[g]indolyl)benzene](/img/es/prev.gif)
![Un estudio cuantitativo de la relación estructura-actividad de una serie de N-[2-(dimetilamino)etil]acridina-4-carboxamidas con actividad citotóxica](/img/es/next.gif)








