Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO
Share
Revista mexicana de ingeniería biomédica
On-line version ISSN 2395-9126Print version ISSN 0188-9532
Rev. mex. ing. bioméd vol.36 n.2 México May./Aug. 2015
https://doi.org/10.17488/RMIB.36.2.4pdf
Artículo de investigación original
Automatic Segmentation of the Cerebellum in Ultrasound Volumes of the Fetal Brain
Segmentación automática del cerebelo en volúmenes de ultrasonido del cerebro fetal
G. Velásquez Rodríguez1, F. Arámbula Cosío1, M.E. Guzmán Huerta2, L. Camargo Marín2, H. Borboa Olivares2, Boris Escalante Ramírez3
1 Laboratorio de Imagenología Biomédica, Centro de Ciencias Aplicadas y Desarrollo Tecnológico, Universidad Nacional Autónoma de México (UNAM).
2 Instituto Nacional de Perinatología, Ciudad de México.
3 Departamento de Procesamiento de Señales, Facultad de Ingeniería, Universidad Nacional Autónoma de México (UNAM).
Correspondencia:
Fernando Arámbula Cosío
Correo electrónico: fernando.arambula@ccadet.unam.mx
Fecha de recepción: 15 de enero de 2015.
Fecha de aceptación: 20 de abril de 2015.
Abstract
The size of the cerebellum in ultrasound volumes of the fetal brain has shown a high correlation with gestational age, which makes it a valuable feature to detect fetal growth restrictions. Manual annotation of the 3D surface of the cerebellum in an ultrasound volume is a time consuming task, which needs to be performed by a highly trained expert. In order to assist the experts in the evaluation of cerebellar dimensions, we developed an automatic scheme for the segmentation of the 3D surface of the cerebellum in ultrasound volumes, using a spherical harmonics model. In this work we present our validation results on 10 ultrasound volumes in which we have obtained an adequate accuracy in the segmentation of the cerebellum (mean Dice coefficient of 0.689). The method reported shows potential to effectively assist the experts in the assessment of fetal growth in ultrasound volumes.
Keywords: 3D fetal ultrasound segmentation, fetal cerebellum, statistical shape models, spherical harmonics.
Resumen
El tamaño del cerebelo, en un volumen de ultrasonido del cerebro fetal, ha mostrado una alta correlación con la edad gestacional, lo que hace importante a esta medición para la detección de restricciones del crecimiento del feto. La anotación manual de la superficie 3D del cerebelo en un volumen de ultrasonido es una tarea demandante, que debe ser realizada por un experto. Con el propósito de apoyar a los expertos en la evaluación de las dimensiones del cerebelo fetal, hemos desarrollado un método automático para la segmentación de la superficie 3D del cerebelo en volúmenes de ultrasonido, utilizando un modelo de harmónicos esféricos (spherical harmonics). En este trabajo presentamos los resultados de una evaluación del método automático en 10 volúmenes de ultrasonido con los que hemos obtenido un valor adecuado de exactitud (coeficiente promedio de Dice de 0.689). El método reportado tiene potencial para asistir de manera efectiva a los expertos en la evaluación del crecimiento fetal, utilizando volúmenes de ultrasonido.
Palabras clave: segmentación 3D de ultrasonido fetal, cerebelo fetal, modelos estadísticos de formas, harmónicos esféricos.
INTRODUCTION
Cerebellar volumes obtained by multiplanar, and the Virtual Organ Computer Aided Analysis (VOCAL) techniques have shown a high correlation with gestational age and might have statistically significant differences between different ethnic groups [1]. Cerebellar volumes evaluated by multiplanar and VOCAL techniques have shown differences between growth restricted fetuses and also adequacy for the assessment of the gestational age [2]. In [3], and [4] is evaluated the growth of the cerebellum in normal pregnancy, using the multiplanar technique. Both techniques (VOCAL and multiplanar evaluation) are useful for fetal evaluation of brain structures, however both have the necessity of manual delineation by an experienced operator, which is considered a disadvantage respect to automated segmentation techniques due to the low reproducibility and agreement obtained by manual segmentation techniques. In order to ease the task for the experts and, more importantly, to contribute to improved reproducibility in the measurement of the cerebellar volume, we have developed two different schemes for automatic segmentation of the cerebellum in ultrasound volumes.
Previous work on the automatic segmentation of fetal brain structures has been reported in Yaqub et al., [12] were a random decision forest classifier was trained to automatically segment: the choroid plexus, the lateral posterior ventricle cavity, the cavum septum pellucidi, and the cerebellum in 3D ultrasound. A mean Dice coefficient of 0.63 was reported for the segmentation of the cerebellum.
In Gutierrez et al., [5] was reported the development of a point distribution model (PDM) of the cerebellum which is automatically adjusted to an ultrasound volume. The model is able to automatically segment the 3D surface of the cerebellum with good accuracy (mean Dice coefficient of 0.8) with a mean processing time of 50 s. Although good results have been achieved with the PDM, in this work we explore the use of a spherical harmonic (SPHARM) model of the cerebellum for automatic segmentation of the structure in ultrasound volumes. SPHARMs have methodological advantages for the construction of statistical deformable models of 3D shapes: There is no need for extensive annotation of landmark points; accurate shape modeling is achieved through principal components analysis of a set of normalized 3D shape examples. In the rest of this work is reported the development of our SPHARM of the cerebellum and its use for the automatic segmentation of the structure in ultrasound volumes. In section 2 we report the construction of the SPHARM of the cerebellum trained from a set of 9 annotated ultrasound volumes. In section 3 we describe the method used to automatically adjust the SPHARM model to an ultrasound volume. In section 4 are reported the results from automatic adjustment of the SPHARM of the cerebellum to a set of 10 different non-training ultrasound volumes, using the leave-one-out method. In section 5 we present the discussion and conclusions of the work reported.
SPHARM MODEL OF THE CEREBELLUM
Our spherical harmonics (SPHARM) model of the cerebellum was trained with 9 cerebellum shapes annotated by an expert in the corresponding ultrasound volumes. Each shape was discretized as show in Fig. 1.

The corresponding triangular mesh was generated for each shape using marching cubes [6]. An example of a mesh is shown in Fig.2.
In order to build the SPHARM model, each mesh of the training see is modeled with spherical harmonic functions as follows [7]:
Spherical harmonic functions of order m and degree l: Ylm, —l ≤ m ≤ l are defined in θ ∈ [0,π], φ ∈ [0, 2π) as:
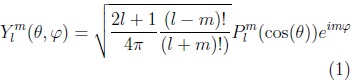
where: plm is the associated Legendre polynomial:
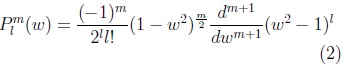
where: w is cos(θ).
To define a surface three SPHARM functions are used, one for each coordinate of the points in the surface:

The surface is then described by the vector-valued function:
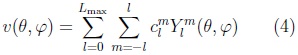
where the coefficients are 3D vectors corresponding to the 3 coordinate functions in (3).
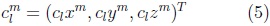
The coefficients are obtained using a least squares adjustment to a given shape mesh, as described in [7].
Using its decomposition in spherical harmonics we can model the 3D surface corresponding to each cerebellum volume in the training set. In Fig. 3 is illustrated an SPHARM model of one cerebellum, we can observe how shape details increase with the degree (Lmax) of the SPHARM.
Once we have a set of SPHARM coefficients for each shape of the cerebellum training set, a principal component analysis (PCA) of the shape parameters is performed through registration of all SPHARM models, using a few corresponding landmark points on each shape [7], for the cerebellum we used 6 landmark points. The Iterative Closest Point method [11] is used to register the shapes in Euclidean space, followed by registration in parameter space through minimization of the Root Mean Square Distance (RMSD) [7]:
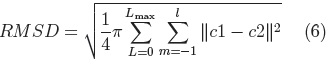
where: c1 and c2 are two SPHARM parameterizations to be registered.
Principal Component Analysis is performed on the parameters (ci, eq.5) of all the examples in the registered training set, to calculate the mean shape and the main modes of shape variation. From which it is possible to generate new shapes of the class of the training set (i.e. cerebellums) using Eq. 7.
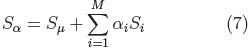
where: Sμ is the mean shape.
Si, are the principal component vectors of the training set and αi are the shape adjustment parameters, with each αi within the range:

λi is the corresponding eigenvalue of each principal component Si.
In this work we used the Matlab toolbox SPHARM-MAT [7] to construct the SPHARM model of the cerebellum. The toolbox generates the parameterizations of each shape on a training set, performs the registration and calculates the mean shape and the main modes of variation (principal components of the covariance matrix). In Fig. 4 is shown our SPHARM model of the cerebellum.
Our SPHARM model was used to automatically segment the surface of the cerebellum in fetal ultrasound volumes, as described in the following section.
AUTOMATIC ADJUSTMENT OF THE SPHARM MODEL TO THE CEREBELLUM IN AN ULTRASOUND VOLUME
We used the method reported by Ahmadi et al [8] for the automatic adjustment of our SPHARM model to the fetal cerebellum in an ultrasound volume. The method is based on the optimization of the following local (gray level) energy function, through gradient descent.

where:
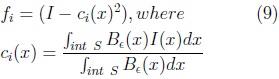

Be is an sphere of radius ∈ used to calculate the values ci and ce for each position x of I. The function E(S) is minimum at the surface which separates two homogeneous regions with significantly different mean grey levels. The gradient of E(S) is given by Eq. 11.
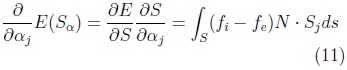
where: N is surface normal and Sj the vertex-wise cartesian deformations for the j-th shape parameter.
Together, these partial derivatives yield the gradient of E with respect to the shape vector a, which we denote by ∇αE .The corresponding discrete equation is:

where: k denotes the vertex number and [.]k denotes the evaluation at vertex k.
Eq. 12 is evaluated for all the vertices of the mesh to find optimum values of the shape parameters (αi) -which accurately fit the cerebellum in an ultrasound volume- through gradient descent.
TESTS AND RESULTS
We used 10 different ultrasound volumes acquired in an axial plane using a Voluson 730 Expert from General Electric, with a 4-8 MHz 3D probe. All volumes were acquired with informed consent of the patients at the National Institute of Perinatology in México City. The cerebellum was annotated on 10 corresponding volumes by an expert sonographer.
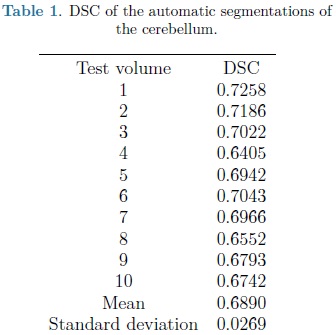
The automatic segmentation of the cerebellum was evaluated on the training set using the leave-one-out method. Nine volumes were used for training the SPHARM of the cerebellum with validation of automatic segmentation on the volume left out. This was repeated 10 times, validating the automatic segmentation on each of the 10 fetal ultrasound volumes. The Dice Similarity Coefficient (DSC) [9] was used to measure the accuracy of the automatic SPHARM segmentation as compared against manual expert annotations.
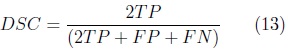
where: TP corresponds to the number of true cerebellum voxels,
FP corresponds to the number of voxels wrongly included as cerebellum and
FN corresponds to the number of cerebellum voxels wrongly left out of the segmentation.
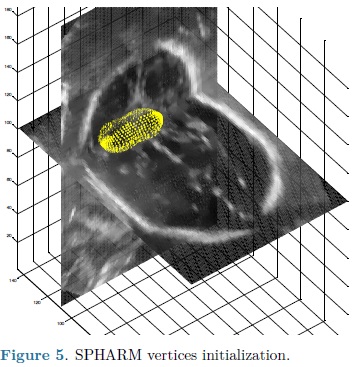
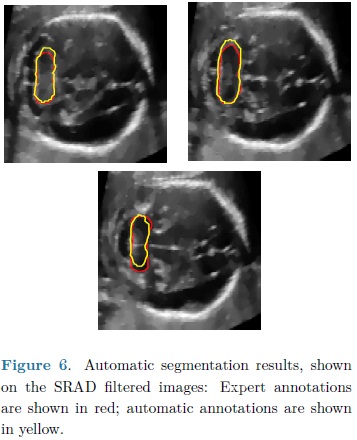
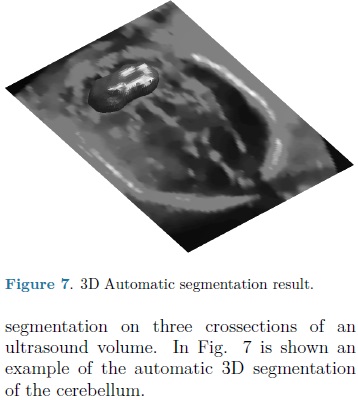
For each automatic segmentation test, the SPHARM was initialized with the mean cerebellum shape and size, located within the central plane of the cerebellum in an ultrasound volume, as shown in Fig. 5. The optimization parameters were: step size of 0.05 for gradient descent, sphere radius of 5 voxels and 30 iterations. Speckle reduction anisotropic diffusion filtering (SRAD) was applied to all ultrasound volumes [10] with 20 iterations and a diffusion coefficient of 0.75. In table 1 are shown the DSC coefficients for our 10 validation ultrasound volumes. In Fig. 6 are shown the results of automatic segmentation on three crossections of an ultrasound volume. In Fig. 7 is shown an example of the automatic 3D segmentation of the cerebellum.
DISCUSSION AND CONCLUSIONS
From table 1 it can be observed that our method is able to automatically segment the cerebellum in an ultrasound volume with adequate accuracy (mean DSC of 0.689). DSC values starting from 0.7 are considered in good agreement with the corresponding expert annotations. In Yaqub et al., [12] a mean DSC of 0.63 was reported for the segmentation of the cerebellum. This, lower, segmentation accuracy is most likely due to the lack of shape models [12].
However higher DSC values have been previously obtained with an automatically adjusted point distribution model (PDM) of the cerebellum, as reported in [5] where 20 ultrasound volumes were used for training and validation using leave-one-out, with a mean DSC for automatic segmentation of 0.8. The gains in accuracy are likely to be due to an improved objective function constructed with grey level voxel profiles, normal to the deformable model surface. In the following stage of development we will explore the use of this objective function with an SPHARM model for automatic segmentation of the cerebellum.
ACKNOWLEDGEMENTS
The authors are grateful to the Mexican National Science and Technology Council (CONACYT) and to the former Institute of Science and Technology of the Federal District in México City, for their financial support to the Biomedical Imaging Lab. at CCADET, UNAM. Gustavo Velásquez is grateful to CONACYT, for the financial support in the form of a PhD scholarship.
REFERENCES
1. E. A. Júnior, H.A. Guimaráes Filho, C.R. Pires, L.M. Nardozza, A.F. Moron, & R. Mattar, "Validation of fetal cerebellar volume by threedimensional ultrasonography in Brazilian population," Archives of Gynecology and Obstetrics, vol. 275, no. 1, pp. 5-11, 2007. [ Links ]
2. A. Benavides-Serralde, E. Hernández-Andrade, J. Fernández-Delgado, W. Plasencia, M. Scheier, F. Crispi, ... & E. Gratacos, "Threedimensional sonographic calculation of the volume of intracranial structures in growth-restricted and appropriate for gestational age fetuses," Ultrasound in Obstetrics & Gynecology, vol. 33, no. 5, pp. 530-537, 2009. [ Links ]
3. T. Hata, A. Kuno, S.Y. Dai, E. Inubashiri, U. Hanaoka, K. Kanenishi, ... & T. Yanagihara, "Threedimensional sonographic volume measurement of the fetal spleen," Journal of Obstetrics and Gynecology Research, vol. 33, no. 5, pp. 600-605, 2007. [ Links ]
4. C.H. Chang, C.H. Yu, F.M. Chang, H.C. Ko, & H.Y. Chen, "The assessment of normal fetal brain volume by 3-D ultrasound," Ultrasound in Medicine & Biology, vol. 29, no. 9, pp. 1267-1272, 2003. [ Links ]
5. B. Gutiérrez-Becker, F. Arámbula Cosío, M.E. Guzmán Huerta, J.A. Benavides-Serralde, L. Camargo-Marín, & V. Medina Bañuelos, "Automatic segmentation of the fetal cerebellum in ultrasound volumes, using a 3D statistical shape model," Medical & Biological Engineering & Computing, vol. 51, no. 9, pp. 1021-1030, 2013. [ Links ]
6. W.E. Lorensen, & H.E. Cline, "Marching cubes: A high resolution 3D surface construction algorithm," ACM Siggraph Computer Graphics, vol. 21, no. 4, pp. 163-169, 1987. [ Links ]
7. L. Shen, H. Farid, & M.A. McPeek, "Modeling threedimensional morphological structures using spherical harmonics," Evolution, vol. 63, no. 4, pp. 1003-1016, 2009. [ Links ]
8. S.A. Ahmadi, M. Baust, A. Karamalis, A. Plate, K. Boetzel, T. Klein, & N. Navab, "Midbrain segmentation in transcranial 3d ultrasound for Parkinson diagnosis," In Medical Image Computing and Computer-Assisted Intervention-MICCAI 2011, pp. 362-369. Springer Berlin Heidelberg, 2011. [ Links ]
9. L.R. Dice, "Measures of the amount of ecologic association between species," Ecology, vol. 26, no. 3, pp. 297-302, 1945. [ Links ]
10. Y. Yu, J.A. Molloy, & S.T. Acton, "Threedimensional speckle reducing anisotropic diffusion," In Signals, Systems and Computers, 2004. Conference Record of the Thirty-Seventh Asilomar Conference on, vol. 2, pp. 1987-1991, IEEE, 2003. [ Links ]
11. P.J. Besl, N.D. McKay, "A method for registration of 3-D Shapes," IEEE Trans. on Pattern Analysis and Machine Intelligence, vol. 14, no. 2, pp. 239-256, 1992. [ Links ]
12. M. Yaqub, R. Cuignet, R. Napolitano, D. Roundhill, A. Papageorghiou, R. Ardon, J.A. Noble, "Volumetric segmentation of key fetal brain structures in 3D ultrasound," In Machine Learning in Medical Imaging 2013, LNCS 8184, pp. 25-32, 2013. [ Links ]














