Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Similares em
SciELO
Similares em
SciELO
Compartilhar
Hidrobiológica
versão impressa ISSN 0188-8897
Hidrobiológica vol.15 no.2 Ciudad de México 2005
Artículo
Structure of the mitochondrial control region and flanking tRNA genes of Mugil cephalus
Estructura de la región control mitocondrial y genes ARNt adyacentes de Mugil cephalus
Axayácatl Rocha-Olivares1, Nikola M. Garber2, Amber F. Garber3, and Kenneth C. Stuck4
1 Department of Biological Oceanography, CICESE, km 107 Carretera Tijuana-Ensenada, Ensenada B. C. 22860.
2 NOAA Sea Grant, 1315 East West Highway, R/SG, Rm 11718, Silver Spring, Maryland 20910.
3 North Carolina State University, Department of Zoology, Campus Box 7617, Raleigh, North Carolina 27695-7617.
4 Gulf Coast Research Laboratory, University of Southern Mississippi, PO Box 7000, Ocean Springs, Mississippi 39566-7000 Dr. Axayácatl Rocha-Olivares CICESE-Department of Biological Oceanography Tel: (646) 175-0500 x 24240 Fax: (646) 175-0545. E. mail: arocha@cicese.mx
Recibido: 4 de octubre 2004.
Aceptado: 27 de mayo 2005.
Abstract
We cloned and sequenced the mitochondrial (mt) control region (CR) and flanking transfer RNA genes (T, P, and F) of the striped mullet, Mugil cephalus and designed species-specific primers to amplify the entire CR in specimens from the Pacific (Hawaii), the Gulf of Mexico, and the Atlantic. We verified the absence of heteroplasmy and nuclear mtDNA duplications of this region in the organisms sampled, finding an extraordinary level of sequence divergence (mean 38-75% Tamura & Nei distance-Γ) between fish from both Oceans, including Japan. The CR of the mullet was variable in length (845-930 bp) and contained structural elements in common with other CRs, including a central conserved segment flanked by hypervariable regions and smaller conserved sequence blocks. Termination associated sequences, however, were not found. The CR of the striped mullet was rich in AT (~67%) and poor in GC.
Key words: Mitochondrial DNA, cloning, molecular divergence, mullet, control region.
Resumen
Se clonó y secuenció la región control (RC) mitocondrial (mt) y los genes de ARNt adyacentes (T, P, y F) de la lisa rayada, Mugil cephalus y se diseñaron cebadores especie-específicos para amplificar la RC en su totalidad en organismos del Pacífico (Hawai), el Golfo de México y el Atlántico. Se verificó la ausencia de heteroplasmia y de duplicaciones nucleares del ADN mt de esta región en los peces analizados, encontrándose una divergencia genética extraordinaria (38-75% distancia Tamura & Nei-Γ) entre las lisas de ambos océanos, incluyendo a Japón. La longitud de la RC fue variable en la lisa (845-930 pb) y presenta elementos estructurales en común con otras RC, incluyendo un segmento central conservado rodeado por dos regiones hipervariables y además bloques de secuencias conservadas más pequeños. No se encontraron secuencias asociadas a la terminación en la RC de la lisa, que se caracterizó por ser rica en AT (~67%) y pobre en GC.
Palabras clave: ADN mitocondrial, clonación, divergencia molecular, lisa, región de control.
Introduction
The vertebrate mitochondrial (mt) genome shows an extreme structural economy with circa 16,000 base pairs (bp) coding for 37 compactly packed genes: two rRNAs, 13 protein open reading frames, and 22 tRNAs (Boore, 1999). Two important structural and functional features of animal mtDNA are the absence of introns (but see Beagley et al., 1998) and of recombination (but see Smith & Smith, 2002). However, the molecule does contain a non-coding region of varying size known as the "control region" (CR), "Displacement-loop containing region", or just "D-loop" (Brown et al., 1986). This non-coding segment contains conserved motifs, the origin of heavy-strand replication, and both heavy- and light-strand transcription initiation sites (Clayton, 1991). In vertebrates, this region is flanked by the genes coding for tRNA-threonine (tRNA-T), tRNA-proline (tRNA-P) at the 5' end of the light strand and for the tRNA-phenylalanine (tRNA-F) at the 3' end (Meyer, 1993). During the evolution of mitochondria from hypothesized -proteobacteria several genes were completely translocated to the eukaryotic nucleus (Gray et al., 1999). Data are accumulating showing mitochondrial duplications (known as nuclear mitochondrial or numt DNA) that have also been copied to the nucleus in several taxa, with the potential confounding and misleading effects on the interpretation of the data if they go unrecognized as pseudogenes (Richly & Leister, 2004).
The rapid evolution of mitochondrial protein coding genes compared to nuclear genes has long been established (Brown et al., 1979) and within the mtDNA, the CR has been estimated to evolve 2-5 times faster than protein coding genes (Meyer, 1993). Due to its elevated evolutionary rate, the CR has been the marker of choice to address a variety of intra-specific genetic questions in a wide range of taxa (v.gr., Taberlet, 1996). Comparative studies of the animal CR have revealed a structure consisting of a central conserved region flanked by two hypervariable sections; as well as the existence of conserved sequence blocks (CSB, < 30 bp), which may be found near the heavy strand origin of replication (Walberg & Clayton, 1981) but also several hundred nucleotides downstream from it (Doda et al., 1981). The latter have not been documented in the species of fish examined to date, although some CSBs are conserved from fish to mammals (v. gr., Lee et al., 1995; Liu et al., 2002). Other structural features of the piscine CR are the termination associated sequences (TAS) (Liu et al., 2002). The 5' hypervariable region of the CR adjacent to tRNA-P is characterized by high levels of nucleotide substitution and, due in part to historical circumstances (since universal primers were first designed for it, Kocher et al., 1989), it has been the most widely used in fish micro- and macroevolutionary genetic research (e.g. Rocha-Olivares et al., 1999a; Rocha-Olivares et al., 1999b; Rocha-Olivares & Vetter, 1999; Rocha-Olivares & Sandoval-Castillo, 2003).
Mugil cephalus Linnaeus 1758, known as striped, grey, or black mullet, or "lisa rayada" (Nelson et al., 2004), is a cosmopolitan species inhabiting tropical and subtropical regions of the world between 42ºN and 42ºS (Gilbert, 1993). The species is of commercial importance in most countries including Mexico (v.gr., Ibañez-Aguirre & Gallardo-Cabello, 1996) and the United States of America (Leber et al., 1996). In part because of the economic relevance, but also because its morphological conservatism and challenging systematics, this species has motivated several genetic studies that have brought to light unsuspected levels of genetic differentiation using biochemical markers and mitochondrial DNA (Tsvetnenko, 1991; Crosetti et al., 1993; Crosetti et al., 1994; Rossi et al., 1998; Rocha-Olivares et al., 2000; Garber et al., 2001; see also Miya et al., 2001 for the complete mitochondrial genome). Here, we report the sequence and structure of the complete CR and flanking tRNA genes of M. cephalus from the Pacific (Hawaii and Japan) and the Atlantic (Gulf of Mexico and Northwest Atlantic) Oceans to establish base line levels of geographic variation in the structure of this important region of the mitochondrial genome in the context of vertebrate control region evolution.
Materials and methods
Mugil cephalus were collected from the Atlantic coast (North Carolina) and the Gulf of Mexico (Florida, Mississippi, Louisiana, and Texas) (n = 96) and from the Pacific Ocean (Island of Oahu, Hawaii) (n = 19), as reported in Rocha-Olivares et al. (2000). Total genomic DNA was isolated by phenol-chloroform extraction from white muscle tissue and quantified using fluorescence spectrophotometry (Sambrook & Russell, 2001).
A segment containing the 3' end of the cytochrome b gene, tRNA-T, tRNA-P, control region, tRNA-F, and the 5' end of the 12S rRNA was amplified by PCR using universal primers CB3 (Palumbi, 1996) and 12SAR (Martin et al., 1992) in replicate 25 µl reactions (100 ng template DNA, 1.5 mM MgCl2, 200 µM dNTPs, 0.3 µM of each primer, 1.75 units of Taq DNA polymerase, 1X PCR buffer Amersham Life Science). Cycling parameters were 3 min at 94ºC, followed by 35 cycles of 45 sec at 94ºC, 1 min at 55ºC, and 2 min at 72ºC, with a final elongation of 7 min at 72ºC. After visualization on a 1% agarose gel, the appropriate PCR product was excised, purified using the QIAquick Gel Extraction Kit (QIAGEN, Inc.), quantified, and direct sequenced. Species specific primers (MulPro and Mul12S) were designed from this product (Fig. 1).
PCR products obtained with species-specific primers were purified and cloned using the pGEM®-T Easy Vector System (Promega, Inc.). Ligated vector DNA was transformed into competent JM109 cells that were cultured on Luria-Bertani (LB)/ampicillin plates with x-gal and IPTG. Colonies containing inserts were identified by blue/white selection and used to inoculate 5 ml minipreps. The cloned plasmid DNA was isolated using the Wizard® Plus DNA Purification System (Promega, Inc.). Plasmid DNA was then purified using PEG (Nicoletti & Condorelli, 1993), quantified, and sequenced.
A species-specific primer in the tRNA-F (MulPhe) was subsequently designed and used with the primer MulPro to amplify the entire control region (Fig. 1) in 50 µL reactions (1x PCR buffer, 200 mM dNTPs, 1.5 µM MgCl2, 0.3 µM of each primer, 200 ng template, and 3.5 units Taq DNA polymerase) with the above PCR cycling parameters. The appropriate PCR-product was gel-purified, quantified, and direct sequenced. All DNA sequencing was completed with an ABI model 373A Stretch Automated DNA Sequencer at the University of Maine DNA Sequencing Facility.
Because of potential of numt duplications, we corroborated the mitochondrial nature of the source DNA conducting nested PCR with three sets of primers previously used to amplify mtDNA sequences. Following amplification with CB3 and 12SAR PCR products (Fig. 1) were visualized on a 1% agarose gel and the appropriate band excised, gel purified, and quantified. This product was then used as template in subsequent nested PCR reactions with three sets of mtDNA CR primers: L15998-PRO TACCCCAAACTCCCAAAGCTA and H00585-PHE CAGTGTTAAGCTTTAACTAAGCT (Alvarado Bremer et al., 1995); "A" TTCCACCTCTAACTCCCAAAGCTAG and "E" CCT-GAAGTAGGAACCAGATG; and "F" CGTCGGATCCAGAGCCTAC-CACAAGGTGATT and "G" CGTCGGATCCCATCTTCAGTGTTATGCTT (Lee et al., 1995).
DNA sequences were aligned using CLUSTAL-W with the default settings and verified by eye. Sequences from the Pacific and Atlantic were used to construct a majority-rule consensus sequence. We used the tRNAscan SE search server to infer the secondary structure of the mitochondrial tRNA genes (Lowe & Eddy, 1997). The M. cephalus sequence used for tRNA analyses was deposited in GenBank accession number AF108270. Intraspecific nucleotide variability in the control region was assessed from all sequence data available that included sequences previously published by us AF108232-352 (Rocha-Olivares et al., 2000), as well as from the complete mitochondrial genome of M. cephalus (accession NC003182, Miya et al., 2001). These data were used to produce a new updated multiple alignment different from that used in Rocha-Olivares et al. (2000). Sequence divergence was estimated accounting for different rates of transition and transversion, unequal base frequencies and among site rate variation with a gamma corrected (shape parameter = 0.5) Tamura and Nei model (TrN-Γ), conceived to model the evolution of the vertebrate control region (Tamura & Nei, 1993). Phylogenetic relationships among haplotypes were reconstructed using the Neighbor-Joining method (Saitou & Nei, 1987) with the program Mega 2.0 (Kumar et al., 2001).
Results
Mullet DNA from Mississippi and Hawaii amplified with the universal primers CB3 and 12SAR resulted in a PCR product ~2000 bp in length. Sequences obtained from clones of a 1300 bp PCR product from this fragment (using MulPro-Mul12S) were identical for a single amplification, indicating the absence of heteroplasmy in the amplified mtDNA. Except for one pair of primers, nested amplifications within CR primer pairs known to be contained within the fragment produced bands of the expected sizes, suggesting that the priming regions have not experienced run away divergent mutation as expected in a pseudogene. Further corroboration of the authenticity of the mtDNA was obtained from NCBI Blast searches in which the sequences were highly homologous to other teleost CRs and particularly to the CR sequence of Mugil cephalus obtained from the entire mitochondrial genome (NC003182).
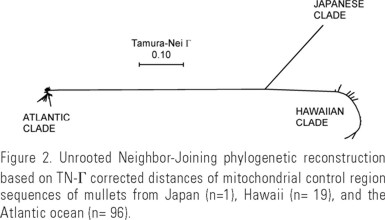
Species-specific primers (MulPro and MulPhe) designed from the cloned fragment (Genbank accession AF108270) produced a fragment ~880 bp in length. M. cephalus control region of the Atlantic specimens (n = 96) ranged in size from 884 - 894 bp (845 bp in one specimen); whereas in the Pacific (Hawaii, n = 19) it ranged from 919-930 bp. All individual sequences were distinct from each other, yielding a total of 115 control region haplotypes. The most relevant finding from the sequencing experiments was the great and unexpected level of DNA divergence found between the Pacific and Atlanic mullets in excess of 23% uncorrected sequence divergence and 70% TrN-Γ corrected divergence (Table 1). This degree of divergence was in sharp contrast with the intra-regional level (Table 1) and resulted in a phylogenetic tree with extremely long branches leading to each cluster of regional haplotypes (Fig. 2). The tree revealed the closer relationship of the Japanese sequence to the Hawaiian haplotypes, despite a considerable level of divergence, and reflected the extreme divergence between mullet from the Pacific and Atlantic oceans (Fig. 2, Table 1).
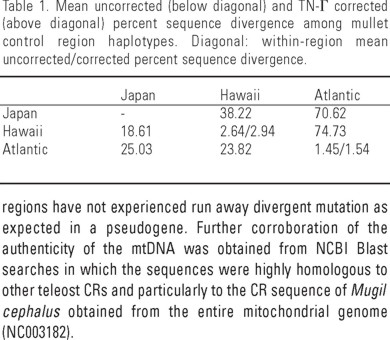
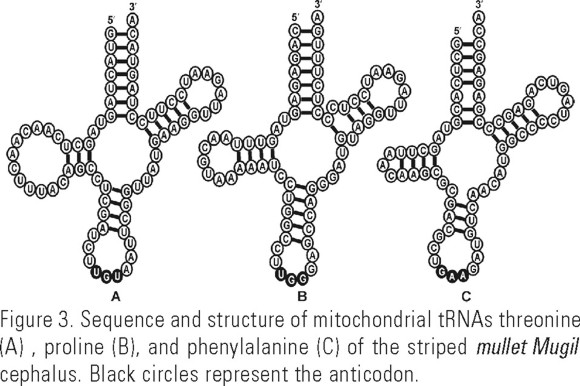
The sequences of the T, P and F tRNA genes were identified in the cloned DNA sequence and the predicted secondary structure resulted in the expected canonical structure of these molecules (Fig. 3). The multiple alignment of sequences from the two ocean basins (Fig. 4), revealed a conserved central portion flanked by the hypervariable regions, with the 5' end of the control region being more variable. CSB-1, 2 and 3, found in most vertebrate control regions, were present in both the Atlantic and Pacific sequences. In the Pacific we also found a putative CSB-D (Fig. 4). Various insertions and deletions were detected, generally associated to repetitive sequences. Termination associated sequence (TAS) regions were not observed in the CR of M. cephalus but the AT-rich 3' end of the control region featured an [AATATTAT] repetitive motif found in several Pacific sequences (from position 984 in Fig. 4). Different direct and indirect repeated elements of eight or greater nucleotides were identified in all fish. Two direct and five indirect and palindromic repeats were observed in Atlantic specimens; whereas nine direct repeats and two indirect and palindromic sequences were found in the Pacific specimens (Table 2). The nucleotide composition of M. cephalus CR was found to be AT rich and poor in G, percent compositions varied from 35.9 -38.1% (T), 34.5 - 38.1% (A), 17.6 - 20.4% (C), and 12.3 - 15.2 % (G), with a ratio AT:CG varying between 2.07 - 2.15.
Discussion
Numt pseudogenes are paralogous mtDNA segments duplicated and transferred to the nuclear genome which can be mistaken for orthologous regions of mtDNA (Lopez et al., 1994). The occurrence of nuclear pseudogenes is not uncommon, and has been characterized in many taxa, including marine species (e.g. Zhang & Hewitt, 1996; Schneider-Broussard & Neigel, 1997). Since a PCR reaction may amplify the target mtDNA region (e.g., the CR), a pseudogene, or both, steps must be taken to verify the amplification of the target mtDNA. One method of verification is via "long PCR" (Cheng et al., 1994), in which a long piece of DNA is amplified and reamplified with nested primers. The initial amplification product of interest is first gel purified to be used as a template for further amplifications with internal primers. The successful nested amplification of several shorter pieces of DNA of the expected size is a strong indication that the original sequence amplified was of mitochondrial origin, since a pseudogene is more likely to have mutated in several internal priming sites and may not be amplifiable.
Dowling et al. (1996) suggested a second verification technique involving enrichment of mtDNA by differential ultra-centrifugation from mitochondria-rich tissues. This decreases the chance of pseudogene amplification but the method is not free of numt contamination. Therefore, the nested PCR approach of long products appears to be more reliable and cost effective. Because two of the three nested primer pairs produced bands of the expected size and because the sequences produced were homologous to a large number of fish CR sequences retrieved from BLAST searches, we conclude that amplified DNA used in this experiment was of mitochondrial origin.
The length of the M. cephalus control region in this study (845-930 bp) fell within the range of sizes found for many other fishes (varying from 804 to 1,500 bp and averaging 990 bp) such as the Gadids (e.g. pollock, haddock, and tomcod, with regions of 868, 856, and 853 bp, respectively) (Lee et al., 1995); common snook, Centropomus undecimalis, 804 bp (Wilson et al., 1997); swordfish, Xiphias gladius, 842 bp (Alvarado Bremer et al., 1995; Rosel & Block, 1996); white sturgeon, Acipenser transmontanus, 761-1,007 bp (Buroker et al., 1990); Atlantic cod, Gadus morhus, 997 bp (Johansen et al., 1990); rainbow trout, Oncorhynchus mykiss, 1,003 bp (Digby et al., 1992); and salmonids, 1,010-1,028 bp (Shedlock et al., 1992). Several flatfish species have larger control regions (~1,500 bp) due to longer repetitive sequences at the 3' end (Lee et al., 1995).
The structure of the M. cephalus control region is similar to that reported for other fish (Buroker et al., 1990; Johansen et al., 1990; Digby et al., 1992), consisting of a conserved central region flanked by two hypervariable segments. Numerous conserved sequence blocks (CSB) have been found in most fish studied (e.g. Lee et al., 1995). Mugil cephalus from the Atlantic contain CSB-3 and putative CSB-2 and CSB-1, and specimens from the Pacific contain CSB-2, CSB-3, putative CSB-D, and two CSB-1. All CSBs, except CSB-D, are found after nucleotide 572, i.e., toward the 3' end of the control region. The position, order, and sequence of CSB-1, 2, and 3 are similar to those found in swordfish (Rosel & Block, 1996), Atlantic cod (Johansen et al., 1990), common snook (Wilson et al., 1997), and salmonids (Shedlock et al., 1992) (Table 3). CSB-2 and CSB-3 sequences are highly conserved among mammals, an amphibian, and other vertebrates. CSB-1 was identical to, and in the same relative position as, CSB-1 in rainbow trout (Digby et al., 1992) and common snook (Wilson et al., 1997). CSB-1 is much smaller than the 26 bp CSB-1 in mouse (Walberg & Clayton, 1981), but is only one bp smaller than CSB-1 in African clawed frog, Xenopus laevis (Roe et al., 1985) and white sturgeon (Buroker et al., 1990).

Fish from Hawaii contain a second CSB-1 close to the 3' end of the control region. This is similar to the arrangement of CSBs found in the white sturgeon (Brown et al., 1993). The CSB-D, found in the swordfish (Rosel & Block, 1996), the freshwater goby Rhinogobius sp. (Chen et al., 1998), and several pleuronectids (greysole, plaice, yellowtail, and winter flounder) (Lee et al., 1995), was similar, and in the same relative position to the other CSBs found in fish from Hawaii. A number of direct and indirect repeats were found in M. cephalus sequences. Wilson et al. (1997) found 18 repeats within the control region of the common snook. In this species a 39 bp tandem repeat spanned nearly half of the control region. Tandem duplications and repeats have been found in many other fish studies producing mtDNA length polymorphisms in the control region (Billington & Hebert, 1991). Bentzen et al. (1988) observed two or three copies of a 1,500 bp repeat in the American shad. In a subsequent study, they found in heteroplasmic duplications in the 3' end of the north-west Atlantic redfish CR (Bentzen et al., 1998). However, before these variations can be used as genetic markers, the transmission genetics must be understood, as it is currently uncertain whether these heteroplasmic variations are inherited by offspring (Mulligan & Chapman, 1989). However, in the case of mullet cloning experiments, no evidence of mitochondrial heteroplasmy was revealed.
Indels are common in the non-coding CR of fish (Billington & Hebert, 1991). The 42-bp-long deletion found in a fish from Mississippi occurred within the first 146 bases adjacent to the 5' end. This is the region of highest sequence variability. The deletion may have arisen from folding events, replication slippage, splicing, deletion, and/or duplications (Moritz et al., 1987). In the white sturgeon, Buroker et al. (1990) suggested that length variation might be due to misalignment before replication of a 82 bp repeat region. However it has been hypothesized that these length variations would be of minimal consequence to the organisms due to the non-coding nature of the control region (Brown, 1983; Moritz et al., 1987).
Finally, as is the case with mtDNA in general and with CR in particular, the CR nucleotide composition of mullet was AT rich (~67%). Johansen et al. (1990) reported the Atlantic cod control region to be 64% AT. Saccone et al. (1987) mentioned that most ATs are found in the hypervariable regions flanking the conserved central domain. This is also true in invertebrate mtDNAs, v.gr., in Drosophila sp. the control region is known as the A + T-rich region (Avise et al., 1987). The extraordinary level of genetic differentiation among these supposedly intra-specific fish has been discussed elsewhere (Rocha-Olivares et al., 2000) and will not be repeated here. However, it is worth mentioning that the availability of the Japanese sequence from Miya et al. (2001), intermediate between the Hawaiian and Atlantic haplotypes, resulted in a better alignment that, we believe, reflects better the orthology of these divergent sequences and higher estimates of sequence divergence that those previously reported by Rocha-Olivares et al. (2000).
Acknowledgments
We would like to thank the following individuals for their assistance in procuring specimens: Jim Francesconi (North Carolina), Ernst Peebles (Florida), David Ziemann (Hawaii), Lawrence Rozas (Texas), Jay Peterson (Louisiana), and Nate Jordan, Jody Peterson, and Jason Steckler (Mississippi). Funding was provided by the National Marine Fisheries Service in cooperative agreement with the National Oceanic and Atmospheric Administration, U.S. Department of Commerce grant, #NA044676FL. Research was conducted under the U.S. Gulf of Mexico Marine Stock Enhancement Consortium. Funding for the leading author came from CICESE and CONACYT. The manuscript benefited from valuable criticisms and suggestions from F. J. García de León and an anonymous reviewer.
References
ALVARADO BREMER, J.R., A.J. BAKER & J. MEJUTO. 1995. Mitochondrial DNA control region sequences indicate extensive mixing of swordfish (Xiphias gladius) populations in the Atlantic Ocean. Canadian Journal of Fisheries and Aquatic Sciences 52(8): 1720-1732. [ Links ]
AVISE, J.C., J. ARNOLD, R.M. BALL, E. BERMINGHAM, T. LAMB, J.E. NEIGEL, C.A. REEB & N.C. SAUNDERS. 1987. Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Annual Review of Ecology and Systematics 18: 489-522. [ Links ]
BEAGLEY, C.T., R. OKIMOTO & D.R. WOLSTENHOLME. 1998. The mitochondrial genome of the sea anemone Metridium senile (Cnidaria): introns, a paucity of tRNA genes, and a near-standard genetic code. Genetics 148(3): 1091-1108. [ Links ]
BENTZEN, P., W.C. LEGGETT & G.G. BROWN. 1988. Length and restriction site heteroplasmy in the mitochondrial DNA of American shad (Alosa sapidissima). Genetics 118(3): 509-518. [ Links ]
BENTZEN, P., J.M. WRIGHT, L.T. BRYDEN, M. SARGENT & K.C.T. ZWANENBURG. 1998. Tandem repeat polymorphism and heteroplasmy in the mitochondrial control region of redfishes (Sebastes: Scorpaenidae). Journal of Heredity 89(1): 1-7. [ Links ]
BILLINGTON, N. & P.D.N. HEBERT. 1991. Mitochondrial DNA diversity in fishes and its implications for introductions. Canadian Journal of Fisheries and Aquatic Sciences 48(Suppl. 1): 80-94. [ Links ]
BOORE, J.L. 1999. Animal mitochondrial genomes. Nucleic Acids Research 27(8): 1767-1780. [ Links ]
BROWN, G.G., G. GADALETA, G. PEPE, C. SACCONE & E. SBISA. 1986. Structural conservation and variation in the D-loop-containing region of vertebrate mitochondrial DNA. Journal of Molecular Biology 192: 503-511. [ Links ]
BROWN, J.R., A.T. BECKENBACH & M.J. SMITH. 1993. Intraspecific DNA sequence variation of the mitochondrial control region of white sturgeon (Acipenser transmontanus). Molecular Biology and Evolution 10(2): 326-341. [ Links ]
BROWN, W.M. 1983. Evolution of mitochondrial DNA. In: Nei, M. & R. K. Koehn, (Eds.). Evolution of genes and proteins. Sinauer Associates Inc., pp. 62-88. [ Links ]
BROWN, W.M., M. GEORGE, JR. & A.C. WILSON. 1979. Rapid evolution of animal mitochondrial DNA. Proceedings of the National Academy of Sciences of the United States of America 76(4): 1967-1971. [ Links ]
BUROKER, N.E., J.R. BROWN, T.A. GILBERT, P.J. O'HARA, A.T. BECKENBACH, W.K. THOMAS & M.J. SMITH. 1990. Length heteroplasmy of sturgeon mitochondrial DNA: An illegitimate model. Genetics 124(1): 157-163. [ Links ]
CHANG, D.D., R.P. FISHER & D.A. CLAYTON. 1987. Roles for a promoter and RNA processing in the synthesis of mitochondrial displacement-loop strands. Biochimica et Biophysica Acta 909: 85-91. [ Links ]
CHEN, I.S., C.H. HSU, C.F. HUI, K.T. SHAO, P.J. MILLER & L.S. FANG. 1998. Sequence length and variation in the mitochondrial control region of two freshwater gobiid fishes belonging to Rhinogobius (Teleostei: Gobioidei). Journal of Fish Biology 53(1): 179-191. [ Links ]
CHENG, S., C. FOCKLER, W.M. BARNES & R. HIGUCHI. 1994. Effective amplification of long targets from cloned Inserts and human genomic DNA. Proceedings of the National Academy of Sciences of the United States of America 91(12): 5695-5699. [ Links ]
CLAYTON, D.A. 1991. Replication and transcription of vertebrate mitochondrial DNA. Annual Review of Cell Biology 7: 453-478. [ Links ]
CROSETTI, D., W.S. NELSON & J.C. AVISE. 1994. Pronounced genetic structure of mitochondrial DNA among populations of the circum-globally distributed grey mullet (Mugil cephalus). Journal of Fish Biology 44(1): 47-58. [ Links ]
CROSETTI, D., J.C. AVISE, F. PLACIDI, A.R. ROSSI & L. SOLA. 1993. Geographic variability in the grey mullet Mugil cephalus: Preliminary results of mtDNA and chromosome analyses. Aquaculture 111(1-4): 95-101. [ Links ]
DIGBY, T.J., M.W. GRAY & C.B. LAZIER. 1992. Rainbow trout mitochondrial DNA: Sequence and structural characteristics of the non-coding control region and flanking tRNA genes. Gene 118(2): 197-204. [ Links ]
DODA, J.N., C.T. WRIGHT & D.A. CLAYTON. 1981. Elongation of displacement-loop strands in human and mouse mitochondrial DNA is arrested near specific template sequences. Proceedings of the National Academy of Sciences of the United States of America 78(10): 6116-6120. [ Links ]
DOWLING, T.E., C. MORITZ, J.D. PALMER & L.H. RIESEBERG. 1996. Nucleic acids III: Analysis of fragments and restriction sites. In: Hillis, D. M., C. Moritz & B. K. Mable, (Eds.). Molecular systematics. Sinauer Associates, pp. 205-247. [ Links ]
GARBER, N.M., A.F. GARBER, K.C. STUCK & W.D. GRATER. 2001. Mitochondrial control region of striped mullet Mugil cephalus: A tool to restore marine fisheries resources. Proceedings of the Gulf and Caribbean Fisheries Institute 52: 352-359. [ Links ]
GILBERT, C.R. 1993. Geographic distribution of the striped mullet (Mugil cephalus Linnaeus) in the Atlantic and eastern Pacific oceans. Florida Scientist 56(4): 204-210. [ Links ]
GRAY, M.W., G. BURGER & B.F. LANG. 1999. Mitochondrial evolution. Science 283(5407): 1476-1481. [ Links ]
IBAÑEZ-AGUIRRE, A. & M. GALLARDO-CABELLO. 1996. Age determination of the grey mullet Mugil cephalus L and the white mullet Mugil curema V (Pisces: Mugilidae) in Tamiahua Lagoon, Veracruz. Ciencias Marinas 22(3): 329-345. [ Links ]
JOHANSEN, S., P.H. GUDDAL & T. JOHANSEN. 1990. Organization of the mitochondrial genome of Atlantic cod, Gadus morhua. Nucleic Acids Research 18(3): 412-419. [ Links ]
KOCHER, T.D., W.K. THOMAS, A. MEYER, S.V. EDWARDS, S. PÄÄBO, F.X. VILLABLANCA & A.C. WILSON. 1989. Dynamics of mitochondrial DNA evolution in animals: Amplification and sequencing with conserved primers. Proceedings of the National Academy of Sciences of the United States of America 86(16): 6196-6200. [ Links ]
KUMAR, S., K. TAMURA, I.B. JAKOBSEN & M. NEI. 2001. MEGA 2: Molecular Evolutionary Genetic Analysis software. Bioinformatics 17(12): 1244-1245. [ Links ]
LEBER, K.M., S.M. ARCE & D.A. STERRITT. 1996. Marine stock-enhancement potential in nursery habitats of striped mullet, Mugil cephalus, in Hawaii. U. S. Fishery Bulletin 94: 452-471. [ Links ]
LEE, W.J., J. CONROY, W.H. HOWELL & T.D. KOCHER. 1995. Structure and evolution of teleost mitochondrial control regions. Journal of Molecular Evolution 41(1): 54-66. [ Links ]
LIU, H.Z., C.S. TZENG & H.Y. TENG. 2002. Sequence variations in the mitochondrial DNA control region and their implications for the phylogeny of the Cypriniformes. Canadian Journal of Zoology 80(3): 569-581. [ Links ]
LOPEZ, J.V., N. YUHKI, R. MASUDA, W.S. MODI & S.J. O'BRIEN. 1994. Numt, a recent transfer and tandem amplification of mitochondrial DNA to the nuclear genome of the domestic cat. Journal of Molecular Evolution 39(2): 174-190. [ Links ]
LOWE, T.M. & S.R. EDDY. 1997. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Research 25(5): 955-964. [ Links ]
MARTIN, A.P., R. HUMPHREYS & S.R. PALUMBI. 1992. Population structure of the armorhead, Pseudopentaceros wheeleri, in the North Pacific Ocean: application of the polymerase chain reaction to fisheries problems. Canadian Journal of Fisheries and Aquatic Sciences 49(11): 2386-2391. [ Links ]
MEYER, A. 1993. Evolution of mitochondrial DNA in fishes. In: Hochachka, P. W. & T. P. Mommsen, (Eds.). Biochemistry and Molecular Biology of Fishes Vol. 2. Elsevier, pp. 1-38. [ Links ]
MIYA, M., A. KAWAGUCHI & M. NISHIDA. 2001. Mitogenomic exploration of higher teleostean phylogenies: A case study for moderate-scale evolutionary genomics with 38 newly determined complete mitochondrial DNA sequences. Molecular Biology and Evolution 18(11): 1993-2009. [ Links ]
MORITZ, C., T.E. DOWLING & W.M. BROWN. 1987. Evolution of animal mitochondrial DNA: Relevance for population biology and systematics. Annual Review of Ecology and Systematics 18: 269-292. [ Links ]
MULLIGAN, T.J. & R.W. CHAPMAN. 1989. Mitochondrial DNA analysis of Chesapeake Bay white perch, Morone americana. Copeia 89(3): 679-688. [ Links ]
NELSON, J.S., E.J. CROSSMAN, H. ESPINOSA-PÉREZ, L.T. FINDLEY, C.R. GILBERT, R.N. LEA & J.D. WILLIAMS. 2004. Common and Scientific Names of Fishes from the United States, Canada, and Mexico. American Fisheries Society, Bethesda. 386 p. [ Links ]
NICOLETTI, V.G. & D.F. CONDORELLI. 1993. Optimized PEG method for rapid plasmid DNA purification: high yield from "midi-prep." BioTechniques 14(4): 535-536. [ Links ]
PALUMBI, S.R. 1996. Nucleic acids II: The polymerase chain reaction. In: Hillis, D. M., C. Moritz & B. K. Mable, (Eds.). Molecular systematics. Sinauer Associates, Inc., pp. 205-247. [ Links ]
RICHLY, E. & D. LEISTER. 2004. NUMTs in sequenced eukaryotic genomes. Molecular Biology and Evolution 21(6): 1081-1084. [ Links ]
ROCHA-OLIVARES, A. & R.D. VETTER. 1999. Effects of oceanographic circulation on the gene flow, genetic structure, and phylogeography of the rosethorn rockfish (Sebastes helvomaculatus). Canadian Journal of Fisheries and Aquatic Sciences 56(5): 803-813. [ Links ]
ROCHA-OLIVARES, A. & J.R. SANDOVAL-CASTILLO. 2003. Mitochondrial diversity and genetic structure in allopatric populations of Pacific red snapper Lutjanus peru. Ciencias Marinas 29(2): 197-209. [ Links ]
ROCHA-OLIVARES, A., R.H. ROSENBLATT & R.D. VETTER. 1999A. Molecular evolution, systematics, and zoogeography of the rockfish subgenus Sebastomus (Sebastes: Scorpaenidae) based on mitochondrial cytochrome b and control region sequences. Molecular Phylogenetics and Evolution 11(3): 441-458. [ Links ]
ROCHA-OLIVARES, A., R.H. ROSENBLATT & R.D. VETTER. 1999B. Cryptic species of rockfishes (Sebastes: Scorpaenidae) in the Southern Hemisphere inferred from mitochondrial lineages. Journal of Heredity 90(3): 404-411. [ Links ]
ROCHA-OLIVARES, A., N.M. GARBER & K.C. STUCK. 2000. High genetic diversity, large inter-oceanic divergence and historical demography of the striped mullet. Journal of Fish Biology 57(5): 1134-1149. [ Links ]
ROE, B.A., D.-P. MA, R.K. WILSON & J.F.H. WONG. 1985. The complete nucleotide sequence of the Xenopus laevis mitochondrial genome. The Journal of Biological Chemistry 260(17): 9759-9774. [ Links ]
ROSEL, P.E. & B.A. BLOCK. 1996. Mitochondrial control region variability and global population structure in the swordfish, Xiphias gladius. Marine Biology 125(1): 11-22. [ Links ]
ROSSI, A.R., M. CAPULA, D. CROSETTI, L. SOLA & D.E. CAMPTON. 1998. Allozyme variation in global populations of striped mullet, Mugil cephalus (Pisces: Mugilidae). Marine Biology 131(2): 203-212. [ Links ]
SACCONE, C., M. ATTIMONELLI & E. SBISA. 1987. Structural elements highly preserved during the evolution of the D-loop-containing region in vertebrate mitochondrial DNA. Journal of Molecular Evolution 26(3): 205-211. [ Links ]
SAITOU, N. & M. NEI. 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4: 406-425. [ Links ]
SAMBROOK, J. & D. RUSSELL. 2001. Molecular cloning. A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. p. [ Links ]
SCHNEIDER-BROUSSARD, R. & J.E. NEIGEL. 1997. A large-subunit mitochondrial ribosomal DNA sequence translocated to the nuclear genome of two stone crabs (Menippe). Molecular Biology and Evolution 14(2): 156-165. [ Links ]
SHEDLOCK, A.M., J.D. PARKER, D.A. CRISPIN, T.W. PIETSCH & G.C. BURMER. 1992. Evolution of the salmonid mitochondrial control region. Molecular Phylogenetics and Evolution 1(3): 179-192. [ Links ]
SMITH, J.M. & N.H. SMITH. 2002. Recombination in animal mitochondrial DNA. Molecular Biology and Evolution 19(12): 2330-2332. [ Links ]
TABERLET, P. 1996. The use of mitochondrial DNA control region sequencing in conservation genetics. In: Smith, T. B. & R. K. Wayne, (Eds.). Molecular Genetic Approaches in Conservation. Oxford University Press, pp. 125-142. [ Links ]
TAMURA, K. & M. NEI. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution 10(3): 512-526. [ Links ]
TSVETNENKO, Y.B. 1991. Biochemical polymorphism and genetic variability of mullets. Journal of Ichthyology 31(2): 64-75. [ Links ]
WALBERG, M.W. & D.A. CLAYTON. 1981. Sequence and properties of the human KB cell and mouse L cell D-loop regions of mitochondrial DNA. Nucleic Acids Research 9(20): 5411-5421. [ Links ]
WILSON, R.R., JR., K.A. DONALDSON, M.E. FRISCHER & T.B. YOUNG. 1997. Mitochondrial DNA control region of common snook and its prospect for use as a genetic tag. Transactions of the American Fisheries Society 126(4): 594-606. [ Links ]
ZHANG, D.X. & G.M. HEWITT. 1996. Nuclear integrations: challenges for mitochondrial DNA markers. Trends in Ecology & Evolution 11(6): 247-251. [ Links ]














