Introduction
Environmental pollution with metals has become a major threat to human health (Nies, 1999). Arsenic (As) is a highly toxic and carcinogenic metalloid that is widely distributed in the environment as a result of geogenic and anthropogenic activities (Cullen & Reimer, 1989). The most common oxidation states of arsenic in ecosystems are the pentavalent arsenate As[V] and trivalent arsenite As[III], the latter form being the more toxic (Oremland & Stolz, 2003; Rosen, 2002). Despite its toxicity, the ancient and constant exposure of bacteria to arsenic has led to the microbe colonization of arsenic-rich environments throughout the development of metabolism coupled biotransformation processes, i.e. reduction, oxidation and methylation (Bentley & Chasteen, 2002; Muller et al., 2007; Osborne & Erlich, 1976; Rosen, 2002; Silver & Phung, 2005) that affects geochemistry, speciation, and toxicity of this element (Islam et al., 2004; Oremland & Stolz, 2003). Due to the ability of bacteria to metabolize highly toxic arsenic compounds into a less toxic form (Oremland, Stolz & Hollibaugh, 2004; Weeger et al., 1999), the isolation and study of arsenic-resistant bacteria is attractive for the establishment of processes to ameliorate the bioavailability of arsenic in contaminated soil and water. Many reports of arsenic resistant bacterial isolates from soil and water include a diversity of bacterial genera such as Microbacterium, Alcaligenes, Staphylococcus, Pseudomonas, Corynebacterium, Xanthomonas, Acinetobacter, Flavimonas, Micrococcus, Bacillus, Aeromonas, Enterobacter and Agrobacterium (Achour, Cordi, Poupin, Bauda, & Billard, 2010; Goswami et al., 2015; Majumder, Ghosh, Saha, Kole & Sarkar, 2013; Mateos, Ordóñez, Letek & Gil, 2006; Mokashi & Paknikar, 2002; Nagvenkar & Ramaiah, 2010; Novick & Roth, 1968; Osborne & Erlich, 1976; Pepi et al., 2007; Salmassi et al., 2002; Selvi et al., 2014) and some extremophiles (Baker-Austin et al., 2007; Branco, Chung, & Morais, 2008; Bruneel et al., 2003; Chen & Shao, 2009; Gihring, Druschel, McCleskey, Hamers & Banfield, 2001) some of which are proposed for use in bioremediation (Banerjee, Datta, Chattyopadhyay & Sarkar, 2011; Goswami et al., 2015; Mateos, et al., 2006).
In Mexico, metal contaminated environments are widely distributed as a consequence of about five centuries of uninterrupted mining activities. However, the study of bacterial diversity is limited in mining sites. The aim of this study was the isolation and characterization of autochthonous arsenic-resistant bacteria with potential application in the biotransformation of arsenic compounds to less toxic ones.
Materials and methods
Bacterial isolates
Arsenic tolerance was evaluated in 28 bacterial cultures isolated from soil samples collected from an area located in Guadalupe, Zacatecas, Mexico, which has been previously reported as a metal-contaminated site (Santos et al., 2006).
Analysis of arsenic tolerance in bacterial isolates
To test the bacterial tolerance to arsenic, different ranges of arsenite and arsenate concentrations were proven based on the levels of bacterial tolerance previously reported (Abbas et al., 2014; Pepi et al., 2007; Selvi et al., 2014). To establish the bacterial tolerance to arsenate in solid media, the Minimum Inhibitory Concentration (MIC) was determined. Briefly, 28 bacterial isolates were inoculated onto LB agar plates supplemented with increasing concentrations of sodium dihydrogen arsenate (NaH2AsO4) (1 mM to 300 mM), and incubated at 37 °C for 24 h to 48 h to obtain visible bacterial colonies. The lowest concentration of metal salt that prevented growth on the plates was recorded as the MIC. The isolates that showed the highest MICs to sodium dihydrogen arsenate (AsT2, AsT5, AsT15, AsT21, and AsT23) were also analyzed to determine its tolerance to arsenite in solid media by the establishment of MIC at increasing concentrations (1 mM to 27 mM) of sodium arsenite (NaAsO2). Arsenite tolerance in liquid media was determined by assessing the MIC values (concentration at which bacterial growth was fully inhibited by arsenic ions), as follows: overnight cultures were diluted 1:100 into fresh LB media supplemented with increasing concentrations of NaAsO2 (2.0 mM, 4.0 mM, 6.0 mM, 8.0 mM, 10.0 mM, and 12.0 mM) and grown at 37 °C with shaking at 200 rpm for 8 h (time at which the end of exponential growth was reached in cultures), then the optical density was spectroscopically determined at 600 nm (OD600nm). At least two independent experiments were carried out.
Molecular identification of As-tolerant isolates
Genomic DNA from Arsenite Tolerant (AsT) isolates was extracted from stationary-phase cultures according to a protocol previously described for gram-positive bacteria (Cutting & Vander Horn, 1990). Amplification of the 16S rDNA gene was accomplished by PCR with the high-fidelity Platinum Pfx DNA-polymerase (Invitrogen, Carlsbad, CA) following the manufacturer’s recommendations, and using 0.3 µM of the primers f D1 (5’ AGAGTTTGATCCTGGCTCAG 3’ corresponding to E. coli 16S rDNA gene bases 1 to 20) and rP2 (5’ ACGGCTACCTTGTTACGACTT 3’ corresponding to E. coli 16S rDNA gene bases 1487 to 1507) (Weisburg, Barns, Pelletier, & Lane, 1991), and 50 ng of DNA template. PCR was performed for 30 cycles, each consisting of a 30 s denaturation step at 94 °C, a 30 s annealing step at 50.9 °C, and a 1 min extension step at 68 °C. The PCR products were purified with the High Pure PCR Product Purification Kit (Roche, Mannheim, Germany) and directly sequenced with the same primers used in PCR. The 16S rDNA assembled sequences of the AsT isolates were deposited in the GeneBank of the National Center for Biotechnology Information (NCBI) with the following accession numbers: KX866673 (AsT2), KX866674 (AsT5), KX866675 (AsT13), KX866676 (AsT14), KX866677 (AsT15), KX866678 (AsT18), KX866679 (AsT21), KX866681 (AsT23), KX866680 (AsT25), and KT717629 AsT27 (the latter previously accessed as PbT5 for its Pb tolerance). The sequences were compared to the 16S ribosomal RNA sequences database of the GeneBank of the National Center for Biotechnology Information server, and sequence alignment was performed using the BLASTN program with a MEGABLAST algorithm for highly similar sequences and default parameters to determine the sequence identity percentages. The determination of bacterial genus (identity ≥ 97%) and species (identity ≥ 99%) was based on the parameters previously described (Drancourt et al., 2000). The 16S rDNA sequences were subjected to pairwise and multiple sequence alignment using the ClustalW program, and the molecular evolutionary analyses were conducted using MEGA6 (Tamura, Stecher, Peterson, Filipski & Kumar, 2013). The sequences used as references for the construction of the phylogenetic tree and their respective accession numbers (gi) for NCBI database are presented below: (631252148) Acinetobacter lwoffii strain JCM 6840, (645320411) Acinetobacter haemolyticus strain ATCC 17906, (645320413) Acinetobacter johnsonii strain ATCC 17909, (636558862) Bacillus simplex strain LMG 11160, (631251529) Bacillus simplex strain NBRC 15720, (343201410) Bacillus simplex strain DSM 1321, (651343563) Micrococcus luteus strain NCTC 2665, (636560518) Micrococcus yunnanensis strain YIM 65004 and (343205881) Micrococcus endophyticus strain YIM 56238.
Morphological and biochemical characterization of As-tolerant strains
Phenotypic analysis of the AsT isolates was based on colony characteristics, Gram staining and biochemical properties such as motility, H2S acid production, and acetoin production from glucose, use of citrate and mannitol as carbon source, and presence of enzymatic activities such as urease, lysine, and ornithine decarboxylases, tryptophanase, cytochrome oxidase, catalase and amylase (MacFaddin, 1984). The identification was established according to the Bergey’s Manual of Determinative Bacteriology (Holt, Rieg, Sneath, Staley & Williams, 1994).
Antimicrobial susceptibility of As-tolerant bacteria
Bacterial resistance to different antibiotics was tested by the disk diffusion method and interpreted according to manufacturer’s instructions for BBL Antibiotic SensiDiscs (Becton Dickinson Microbiology Systems, Cockeysville, MD) or Multidiscos* (BIO RAD, Mexico).
Results and discussion
Tolerance to arsenate (AsO4)-3 and arsenite (AsO3)3- ions in bacterial strains isolated from metal contaminated soil
Due to the constant exposure to arsenic concentrations in metal-contaminated sites, bacteria have developed adaptive strategies to overcome the toxicity of arsenic. In this work, we found a prevalence of gram-positive and bacilli shape in a group of 28 bacterial strains isolated from contaminated soil with arsenic. This results are consistent with previous reports that indicate a high prevalence of gram-positive species of the Arthrobacter, Bacillus, and Micrococcus genera, although gram-negative genera such as Pseudomonas, Flavobacterium, Acinetobacter, Agrobacterium, and Nocardia can also be found (Alexander, 1980; Anderson & Cook, 2004; Grant, 1982; Jackson, Harrison & Dugas, 2005). The analysis of arsenic tolerance of 28 bacterial isolates revealed that MICs of sodium arsenate (NaH2AsO4) and sodium arsenite (NaAsO2) in LB agar were in a range of 0 mM and 300 mM and 4 mM to 25 mM, respectively (Tables 1 and 2), five of this isolates (AsT2, AsT5, AsT15, AsT21, and AsT23) showed a high tolerance to arsenate and arsenite when compared with those reported in genera such as Exiguobacterium, Aeromonas, Bacillus, Pseudomonas, Escherichia, Acinetobacter with CMIs of 10 mM to 275 mM of sodium arsenate As(IV) and 20 mM of sodium arsenite As(III) (Anderson & Cook, 2004; Jackson et al., 2005).
Table 1 Tolerance of bacterial isolates to sodium arsenate (NaH2AsO4).
| Isolate | MIC NaH2AsO4 (mM) | Isolate | MIC NaH2AsO4 (mM) |
|---|---|---|---|
| AsT-1 | 20 | AsT-15 | 300* |
| AsT-2 | 40* | AsT-16 | 2.5 |
| AsT-3 | 4.5 | AsT-17 | 4.0 |
| AsT-4 | 1.0 | AsT-18 | 300* |
| AsT-5 | 300* | AsT-19 | 4.0 |
| AsT-6 | 5.0 | AsT-20 | 2.0 |
| AsT-7 | 2.5 | AsT-21 | 50* |
| AsT-8 | 20 | AsT-22 | 4.0 |
| AsT-9 | 1.5 | AsT-23 | 300* |
| AsT-10 | 2.5 | AsT-24 | 1.0 |
| AsT-11 | 0 | AsT-25 | 300* |
| AsT-12 | 0 | AsT-26 | 4.0 |
| AsT-13 | 60* | AsT-27 | 40* |
| AsT-14 | 200* | AsT-28 | 4.0 |
(*) Highest MIC strains selected for biochemical and 16s gene sequencing identification and analyses of arsenite-tolerance.
Source: Author’s own elaboration.
Table 2 Tolerance of bacterial isolates to sodium arsenite (NaAsO2).
| Isolate | MIC NaAsO2 (mM) |
|---|---|
| AsT-2 | 8.0* |
| AsT-5 | 8.0* |
| AsT-13 | 4.0 |
| AsT-14 | 7.5 |
| AsT-15 | 25.0* |
| AsT-18 | 5.0 |
| AsT-21 | 13.0* |
| AsT-23 | 15.0* |
| AsT-25 | 5.0 |
| AsT-27 | 7.5 |
*() Isolates with the highest MIC were selected to analyze the effects of arsenite ions (AsO3)3- over bacterial growth in liquid culture.
Source: Author’s own elaboration.
The isolates with MICs ≥ 40 mM of NaH2AsO4 -AsT2 (40 mM), AsT5 (300 mM), AsT13 (60 mM), AsT14 (200 mM), AsT15 (300 mM), AsT18 (300 mM), AsT21 (50 mM), AsT23 (300 mM), AsT25 (300 mM) and AsT27 (40 mM)- were selected to analyze tolerance to arsenite ions (AsO3)3-. The MICs values were determined between 4 and 25 mM of NaAsO2 (Table 2) and isolates with the highest tolerance to both arsenate (AsO4)-3 and arsenite (AsO3)3- ions (AsT2, AsT5, AsT15, AsT21, and AsT23) in solid media, were selected to analyze growth in liquid cultures supplemented with increasing concentrations of sodium arsenite (NaAsO2). The AsT23 isolate, with a MIC value of 12 mM of sodium arsenite, showed a higher tolerance to the toxic effects of arsenite ions (AsO3)3- in liquid media (Figure 1e) with respect to isolates AsT2 (7.5 mM), AsT5 (7.5 mM), AsT15 (8 mM) and AsT21 (10 mM) and those previously reported (Banerjee et al., 2011). MICs values indicate that whereas the toxic effects of arsenite ions (AsO3)3- over bacterial growth in both liquid and solid media were similar in the AsT2 and AsT5, the isolates AsT15, AsT21, and AsT23 were more sensitive to arsenite ions in liquid media than in solid media (Table 2 and Figure 1), particularly, AsT15 showed a significant decrease of arsenite tolerance in liquid (MIC 8.0 mM) when compared with tolerance in solid media (MIC 25 mM) (Table 2 and Figure 1), this results suggest that in the isolates AsT2 and AsT5 the arsenite tolerance is probably related to the presence of an arsenite oxidase (encoded by aso genes) that functions as an initial electron donor in aerobic resistance to arsenite allowing the isolates to grow in liquid media under aerobic conditions. On the other hand, the tolerance of AsT15, AsT21, and AsT23 to arsenite in liquid media probably decreased due to the absence of arsenite oxidase in the isolates, whereupon tolerance may be mediated by alternative arsenic-resistance mechanisms (Silver & Phung, 2005). However, future studies are required to establish the mechanisms involved in arsenic tolerance of the AsT. Thus, the high tolerance observed indicate that the AsT isolates analyzed in this work are potential candidates for the study of molecular mechanisms involved in tolerance/resistance to arsenic, as well as to evaluate the ability to bioaccumulate or biotransform toxic arsenic compounds to less toxic ones.
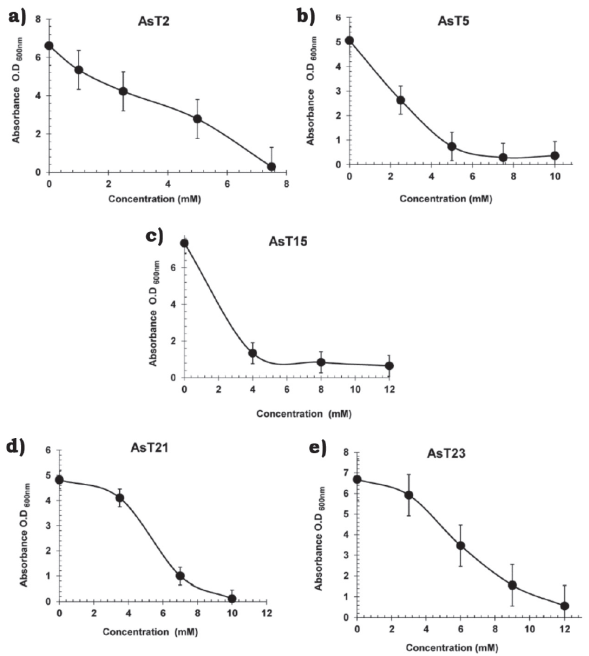
Source: Author’s own elaboration.
Figure 1 Effect of arsenite ions on growth of different bacterial strains exposed to increasing concentrations of NaAsO2 in liquid culture. a) Bacillus simplex, AsT2; b) Micrococcus luteus AsT5; c) Bacillus simplex AsT15; d) Bacillus simplex AsT21; e) Micrococcus sp. AsT23. Overnight cultures were inoculated into fresh LB broth with increasing amounts of NaAsO2 and incubated for 8 h at 37 °C and 200 rpm; the O.D600nm was measured. Data are the average of two independent experiments.
Identification of As-tolerant bacteria such as Bacillus, Micrococcus, and Acinetobacter species
Morphological and biochemical characteristics such as cocci shape, positive Gram staining, non-fermenter of glucose and mannitol, as well as the presence of catalase and oxidase enzymatic activities identified the isolates AsT5 and AsT23 as members of the Micrococcus genus based on the biochemical characteristics reported for this genus (Table 3) (Holt et al., 1994; Kocur, Kloos & Schleifer, 2006). The isolate AsT13 was identified as a member of the Acinetobacter genus based on its non-motil gram-negative cocci shape, aerobic, and non-fermenter metabolism, and the absence of oxidase activity in agreement with characteristics reported for this genus (Hernández Torres, García Vásquez, Yagüe & Gómez Gómez, 2010; Holt et al., 1994) (Table 3). On the other hand, isolates AsT2, AsT14, AsT15, AsT18, AsT21, AsT25 and AsT27 were identified as members of the Bacillus genus based on its bacilli shape, positive gram staining, ability to produce endospores, aerobic facultative growth and the presence of catalase and amylase enzymatic activities (Holt et al., 1994) (Table 3).
Table 3 Morphology and Biochemical characteristics of arsenic-tolerant soil bacteria.
| Morphology | Biochemical properties | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbohydrate metabolism | Enzymatic Activities | |||||||||||||||
| Strain | Colony | Gram stain | Spore former | Motility | H2S production | Acid from glucose | Voges Proskauer | Mannitol | Citrate | Urease | Lysine Descarboxilase | Trypto-phanase | Ornithine descarboxilase | Citochrome Oxidase | Catalase | Amylase |
| AsT-2 (Bacillus simplex) | Yellow, smooth, round, convex. 4 mm - 6 mm diameter | Bacilli Gram positive | + | + | - | + | - | - | + | - | + | - | - | - | + | - |
| AsT-5 (Micrococcus luteus) | Yellow, smooth, round 2 mm - 3 mm diameter | Cocci Gram positive | - | - | - | - | - | - | - | - | + | + | - | + | + | + |
| AsT-13 (Acinetobacter iwofii) | White, creamy, smooth, round, convex 2 mm - 3 mm diameter | Cocci Gram negative | - | - | - | - | - | - | - | - | + | + | - | - | + | - |
| AsT-14 (Bacillus simplex) | Irregular, gray, umbonate, flat 4 mm- 6 mm diameter | Long bacilli Gram positive | + | + | - | + | - | - | - | - | - | - | - | + | + | + |
| AsT-18 (Bacillus simplex) | Irregular, white, umbonate, flat. 4 mm - 6 mm diameter | Long bacilli Gram positive | + | + | - | + | - | - | - | - | - | - | - | - | + | + |
| AsT-21 (Bacillus simplex) | Irregular, white, umbonate, flat 4 mm - 6 mm diameter | Bacilli Gram positive | + | + | - | + | - | - | - | - | + | - | - | + | + | + |
| AsT-23 (Micrococcus sp.) | Punctiform, round, light yellow, smooth 1 mm - 3 mm diameter | Cocci Grampositive | - | - | - | - | - | - | - | - | + | - | - | + | + | + |
| AsT-25 (Bacillus simplex) | Regular, white, flat, creamy, 4 mm - 6 mm diameter | Bacilli Gram positive | + | + | - | + | - | - | - | - | - | - | - | + | + | + |
| AsT-27 (Bacillus simplex) | Regular, cream, flat, shinny, smooth 5 mm - 7 mm diameter | Bacilli Gram positive | + | + | - | + | - | - | + | - | + | - | - | - | + | - |
Source: Author’s own elaboration.
We conducted molecular identification based on amplification and sequence analysis of the 16S rDNA ribosomal gene of the arsenic-tolerant isolates that corroborates with phenotypic identification, this analysis identified the AsT2, AsT14, AsT15, AsT18, AsT21, AsT25, and AsT27 isolates as Bacillus simplex (99% identity), the AsT13 was identified as Acinetobacter lwoffii (99% identity) and the AsT5 and AsT23 isolates as Micrococcus luteus (99% identity) and Micrococcus sp. (89% identity), respectively. The phylogenetic analysis grouped the AsT isolates into three different phyla that include firmicutes (Bacillus species), actinobacteria (Micrococcus species), and proteobacteria (Acinetobacter species) (Figure 2), in accordance with the wide distribution of arsenic resistance in bacteria. The identification showed a high prevalence of the Bacillus genus, which is widely distributed in natural environments (Castillo, Sosa & Scorza, 2004; Felske, Heyrman, Balcaen & De Vos, 2003). Moreover, the results are in agreement with previous studies that report the isolation of bacteria from arsenic contaminated soil that pertain to Bacillus, Pseudomonas, Acinetobacter, Artrobacter, and Micrococcus genus (Archour, Bauda & Billard, 2007; Megharaj, Avudainayagam & Naidu, 2003), the results indicate that arsenic tolerance is widely distributed between these genus in arsenic contaminated environments.
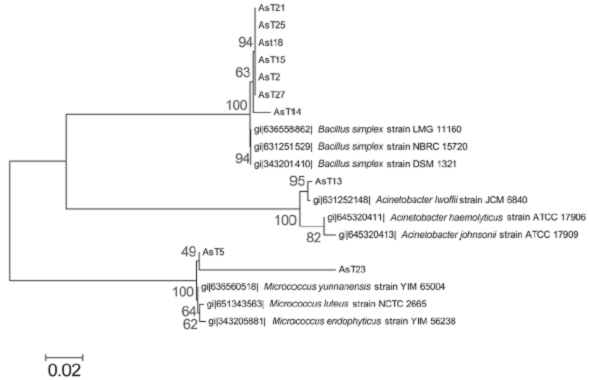
Source: Author’s own elaboration.
Figure 2 Molecular phylogenetic analysis of AsT isolates. The evolutionary history was inferred by using the Maximum Likelihood method based on the Tamura-Nei model (Tamura & Nei, 1993). The tree with the highest log likelihood (-3200.2978) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained by applying the Neighbor-Joining method to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 19 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 961 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 (Tamura et al., 2013).
The identification of the Acinetobacter species is consistent with the normal habitat of the most species of this genus, which is widely distributed in different water and soil environments, even in soils with organic pollutants and heavy metals such as arsenic (Archour et al., 2007; Cai, Liu, Rensing & Wang, 2009). However, some species such as A. baumannii and A. johnsonii, A. haemolyticus and A. lwoffi, also present as a normal part of the skin microbiota, mucous membranes, respiratory secretions, urine and other clinical samples, and are commonly associated with opportunistic infections especially in patients with a compromised immune systems in hospital environments (Barbe et al., 2004; Guardabassi, Dalsgaard & Olsen, 1999; Hernández Torres et al., 2010). On the other hand, it has been reported that members of the Acinetobacter genus have great potential in biotechnological applications, degradation of hydrocarbons in diesel contaminated soils, and bioremediation (Gallego, Loredo, Llamas, Vázquez & Sánchez, 2001; Koma et al., 2001) and Micrococcus luteus has been widely used in biotechnological biodegradation and/or bioremediation processes because it has been shown that this organism is capable of using some contaminants as a food source (Kayode-Isola, Eniola, Olayemi & Igunnugbemi, 2008). Therefore, the results obtained in this work are the basis for future studies that are needed to establish the potential of these arsenic-tolerant isolates to be used in biotechnological processes.
Antibiotic resistance of Arsenic-tolerant bacteria
The analysis of antibiotic resistance revealed a moderate prevalence of resistance (in at least 50% of isolates) to beta-lactamics such as cefotaxime, ceftazidime, cefuroxime, dicloxacillin and penicillin, whereas resistance to antibiotics such as vancomycin (glycopeptide), nitrofurantoin (sulfamide), and ceftriaxone (cephalosporin) was observed in a 30% to 40% of the arsenic-tolerant isolates. In contrast, a low prevalence of resistance (0% to 10% of isolates) to ampicillin, levofloxacin, cephalothin, chloramphenicol, netilmicin, cefepime, sulfamethoxazole, pefloxacin, tetracycline, ciprofloxacin, and erythromycin was observed (Table 4). The isolation of an Acinetobacter lwoffi strain (opportunistic pathogen) resistant to several antibiotics at the sampling site, which is currently urbanized, is indicative of the distribution of multi-resistant pathogens to antibiotics in natural environments, which have also developed a high tolerance and/or resistance to toxic metals. The above result is supported by previous studies that show an association between plasmid-mediated metal tolerance and multiple antibiotic resistance (Calomiris, Armstrong & Seidler, 1984; Tewari, Ramteke, Tripathi, Kumar & Garg, 2013; Timoney, Port, Giles & Spanier, 1998). Therefore, the results contribute to the understanding of the public health problem in Zacatecas related to multi-resistance to antibiotics by human and animal pathogens.
Table 4 Antimicrobial susceptibility of arsenic-tolerant soil bacteria.
| Susceptibility of bacterial isolates to antibiotics | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 a | 2 b | 3 c | 4 c | 5 c | 6 c | 7 c | 8 d | 9 c | 10 c | 11 | 12 b | 13 f | 14 e | 15 c | 16 c | 17 b | 18 c | 19 g | 20 c | h | |
| AsT2 (B. simplex) | S | S | S | R | S | S | S | S | R | R | R | S | R | S | S | S | S | S | S | R | S |
| AsT5 (M. luteus) | S | S | S | S | S | S | S | S | S | R | R | R | R | S | S | S | S | S | S | S | S |
| AsT13 (A. lwoffii) | S | S | R | S | S | S | S | S | S | S | R | S | R | S | S | S | S | R | S | S | R |
| AsT14 (B. simplex) | S | S | S | R | S | S | S | S | R | R | S | S | S | S | S | S | S | S | S | R | S |
| AsT15 (B. simplex) | S | S | S | R | S | S | R | S | R | R | R | S | S | S | S | S | S | R | S | S | S |
| AsT18 (B. simplex) | S | S | S | R | S | S | S | S | R | S | R | S | R | S | R | S | S | S | R | R | S |
| AsT21 (B. simplex) | R | S | S | R | S | S | S | S | R | R | R | S | R | S | S | S | S | S | S | S | S |
| AsT23 (Micrococcus sp.) | S | S | S | S | S | S | S | S | S | S | R | R | S | S | S | S | S | S | S | S | S |
| AsT25 (B. simplex) | S | S | S | S | S | S | S | S | R | R | R | S | R | S | S | S | S | R | S | R | R |
| AsT27 (B. simplex) | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | R | S | S | R |
1: Ampicillin; 2: Levofloxacin; 3: Cephalothin; 4: Cefotaxime; 5: Col; 6: Netilmicin; 7: Cefepime; 8: Trimeth/Sulpha; 9: Ceftazidime; 10: Cefuroxime; 11: Dicloxacillin; 12: Pefloxacine; 13: Penicillin; 14: Tetracyclin; 15: Amikacin; 16: Gentamicin; 17: Ciprofloxacin; 18: Vancomycin; 19: Erythromycin; 20: Ceftriaxone.
R: resistant; S:susceptible. Antibiotic concentrations: a : 10 µg; b : 5 µg; c : 30 µg; d : Trimethoprim/Sulphamethoxazole 25 µg; e : 1 µg; f : 10 IU; g : 15 µg; h : 300 µg. (BBL Antibiotic SensiDiscs, BD. Gram Positive and Gram Negative II Multidisc, BIO-RAD).
Source: Author’s own elaboration.
Conclusions
The identification and characterization of autochthonous bacteria with high tolerance to arsenic conducted in this study constitutes the first approach to investigate the mechanisms involved in arsenic resistance and its relationship with antibiotic multiresistance to contribute to the discovery of soil microbes with potential applications for biotransformation and waste treatment of arsenic-polluted sites.











 nueva página del texto (beta)
nueva página del texto (beta)


